Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin
- PMID: 24803516
- PMCID: PMC4010824
- DOI: 10.1128/mBio.00966-14
Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin
Abstract
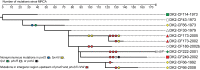
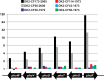
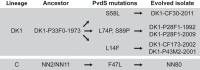
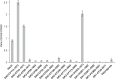
ABSTRACT Pseudomonas aeruginosa airway infections are a major cause of mortality and morbidity of cystic fibrosis (CF) patients. In order to persist, P. aeruginosa depends on acquiring iron from its host, and multiple different iron acquisition systems may be active during infection. This includes the pyoverdine siderophore and the Pseudomonas heme utilization (phu) system. While the regulation and mechanisms of several iron-scavenging systems are well described, it is not clear whether such systems are targets for selection during adaptation of P. aeruginosa to the host environment. Here we investigated the within-host evolution of the transmissible P. aeruginosa DK2 lineage. We found positive selection for promoter mutations leading to increased expression of the phu system. By mimicking conditions of the CF airways in vitro, we experimentally demonstrate that increased expression of phuR confers a growth advantage in the presence of hemoglobin, thus suggesting that P. aeruginosa evolves toward iron acquisition from hemoglobin. To rule out that this adaptive trait is specific to the DK2 lineage, we inspected the genomes of additional P. aeruginosa lineages isolated from CF airways and found similar adaptive evolution in two distinct lineages (DK1 and PA clone C). Furthermore, in all three lineages, phuR promoter mutations coincided with the loss of pyoverdine production, suggesting that within-host adaptation toward heme utilization is triggered by the loss of pyoverdine production. Targeting heme utilization might therefore be a promising strategy for the treatment of P. aeruginosa infections in CF patients. IMPORTANCE Most bacterial pathogens depend on scavenging iron within their hosts, which makes the battle for iron between pathogens and hosts a hallmark of infection. Accordingly, the ability of the opportunistic pathogen Pseudomonas aeruginosa to cause chronic infections in cystic fibrosis (CF) patients also depends on iron-scavenging systems. While the regulation and mechanisms of several such iron-scavenging systems have been well described, not much is known about how the within-host selection pressures act on the pathogens' ability to acquire iron. Here, we investigated the within-host evolution of P. aeruginosa, and we found evidence that P. aeruginosa during long-term infections evolves toward iron acquisition from hemoglobin. This adaptive strategy might be due to a selective loss of other iron-scavenging mechanisms and/or an increase in the availability of hemoglobin at the site of infection. This information is relevant to the design of novel CF therapeutics and the development of models of chronic CF infections.
Figures

References
-
- Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Workman CT, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. U. S. A. 108:7481–7486. 10.1073/pnas.1018249108 - DOI - PMC - PubMed
-
- Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492. 10.1073/pnas.0602138103 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

