Detection of evolutionarily distinct avian influenza a viruses in antarctica
- PMID: 24803521
- PMCID: PMC4010832
- DOI: 10.1128/mBio.01098-14
Detection of evolutionarily distinct avian influenza a viruses in antarctica
Abstract
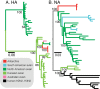
ABSTRACT Distinct lineages of avian influenza viruses (AIVs) are harbored by spatially segregated birds, yet significant surveillance gaps exist around the globe. Virtually nothing is known from the Antarctic. Using virus culture, molecular analysis, full genome sequencing, and serology of samples from Adélie penguins in Antarctica, we confirmed infection by H11N2 subtype AIVs. Their genetic segments were distinct from all known contemporary influenza viruses, including South American AIVs, suggesting spatial separation from other lineages. Only in the matrix and polymerase acidic gene phylogenies did the Antarctic sequences form a sister relationship to South American AIVs, whereas distant phylogenetic relationships were evident in all other gene segments. Interestingly, their neuraminidase genes formed a distant relationship to all avian and human influenza lineages, and the polymerase basic 1 and polymerase acidic formed a sister relationship to the equine H3N8 influenza virus lineage that emerged during 1963 and whose avian origins were previously unknown. We also estimated that each gene segment had diverged for 49 to 80 years from its most closely related sequences, highlighting a significant gap in our AIV knowledge in the region. We also show that the receptor binding properties of the H11N2 viruses are predominantly avian and that they were unable to replicate efficiently in experimentally inoculated ferrets, suggesting their continuous evolution in avian hosts. These findings add substantially to our understanding of both the ecology and the intra- and intercontinental movement of Antarctic AIVs and highlight the potential risk of an incursion of highly pathogenic AIVs into this fragile environment. IMPORTANCE Avian influenza viruses (AIVs) are typically maintained and spread by migratory birds, resulting in the existence of distinctly different viruses around the world. However, AIVs have not previously been detected in Antarctica. In this study, we characterized H11N2 viruses sampled from Adélie penguins from two geographically different sites in Antarctica and show that the segmented AIV genome diverged between 49 and 80 years ago from other AIVs, with several genes showing similarity and shared ancestry with H3N8 equine influenza viruses. This study provides the first insight into the ecology of AIVs in Antarctica and highlights the potential risk of an introduction of highly pathogenic AIVs into the continent.
Figures


References
-
- Vijaykrishna D, Deng YM, Su YC, Fourment M, Iannello P, Arzey GG, Hansbro PM, Arzey KE, Kirkland PD, Warner S, O’Riley K, Barr IG, Smith GJ, Hurt AC. 2013. The recent establishment of North American H10 lineage influenza viruses in Australian wild waterfowl and the evolution of Australian avian influenza viruses. J. Virol. 87:10182–10189. 10.1128/JVI.03437-12 - DOI - PMC - PubMed
-
- Alvarez P, Mattiello R, Rivailler P, Pereda A, Davis CT, Boado L, D’Ambrosio E, Aguirre S, Espinosa C, La Torre J, Donis R, Mattion N. 2010. First isolation of an H1N1 avian influenza virus from wild terrestrial non-migratory birds in Argentina. Virology 396:76–84. 10.1016/j.virol.2009.10.009 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

