Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster
- PMID: 24803674
- PMCID: PMC4066794
- DOI: 10.1093/nar/gku387
Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster
Erratum in
-
Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster.Nucleic Acids Res. 2015 Mar 11;43(5):2991. doi: 10.1093/nar/gkv117. Epub 2015 Feb 11. Nucleic Acids Res. 2015. PMID: 25675957 Free PMC article. No abstract available.
Abstract
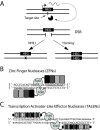
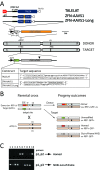
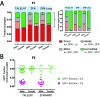
Selfish genes are DNA elements that increase their rate of genetic transmission at the expense of other genes in the genome and can therefore quickly spread within a population. It has been suggested that selfish elements could be exploited to modify the genome of entire populations for medical and ecological applications. Here we report that transcription activator-like effector nuclease (TALEN) and zinc finger nuclease (ZFN) can be engineered into site-specific synthetic selfish elements (SSEs) and demonstrate their transmission of up to 70% in the Drosophila germline. We show here that SSEs can spread via DNA break-induced homologous recombination, a process known as 'homing' similar to that observed for homing endonuclease genes (HEGs), despite their fundamentally different modes of DNA binding and cleavage. We observed that TALEN and ZFN have a reduced capability of secondary homing compared to HEG as their repetitive structure had a negative effect on their genetic stability. The modular architecture of ZFNs and TALENs allows for the rapid design of novel SSEs against specific genomic sequences making them potentially suitable for the genetic engineering of wild-type populations of animals and plants, in applications such as gene replacement or population suppression of pest species.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Burt A., Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Harvard University Press; 2006.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

