Advanced methods for the analysis of chromatin-associated proteins
- PMID: 24803678
- PMCID: PMC4080278
- DOI: 10.1152/physiolgenomics.00041.2014
Advanced methods for the analysis of chromatin-associated proteins
Abstract
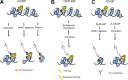
DNA-protein interactions are central to gene expression and chromatin regulation and have become one of the main focus areas of the ENCODE consortium. Advances in mass spectrometry and associated technologies have facilitated studies of these interactions, revealing many novel DNA-interacting proteins and histone posttranslational modifications. Proteins interacting at a single locus or at multiple loci have been targeted in these recent studies, each requiring a separate analytical strategy for isolation and analysis of DNA-protein interactions. The enrichment of target chromatin fractions occurs via a number of methods including immunoprecipitation, affinity purification, and hybridization, with the shared goal of using proteomics approaches as the final readout. The result of this is a number of exciting new tools, with distinct strengths and limitations that can enable highly robust and novel chromatin studies when applied appropriately. The present review compares and contrasts these methods to help the reader distinguish the advantages of each approach.
Keywords: DNA-protein interactions; affinity purification; chromatin; chromatin enrichment; mass spectrometry.
Copyright © 2014 the American Physiological Society.
Figures

References
-
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 12: 123–135, 2011 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

