c-kit+ cells minimally contribute cardiomyocytes to the heart
- PMID: 24805242
- PMCID: PMC4127035
- DOI: 10.1038/nature13309
c-kit+ cells minimally contribute cardiomyocytes to the heart
Abstract
If and how the heart regenerates after an injury event is highly debated. c-kit-expressing cardiac progenitor cells have been reported as the primary source for generation of new myocardium after injury. Here we generated two genetic approaches in mice to examine whether endogenous c-kit(+) cells contribute differentiated cardiomyocytes to the heart during development, with ageing or after injury in adulthood. A complementary DNA encoding either Cre recombinase or a tamoxifen-inducible MerCreMer chimaeric protein was targeted to the Kit locus in mice and then bred with reporter lines to permanently mark cell lineage. Endogenous c-kit(+) cells did produce new cardiomyocytes within the heart, although at a percentage of approximately 0.03 or less, and if a preponderance towards cellular fusion is considered, the percentage falls to below approximately 0.008. By contrast, c-kit(+) cells amply generated cardiac endothelial cells. Thus, endogenous c-kit(+) cells can generate cardiomyocytes within the heart, although probably at a functionally insignificant level.
Conflict of interest statement
None
Figures
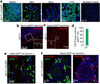

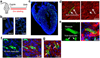

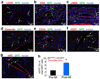


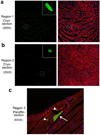


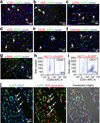

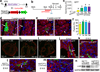
Comment in
-
Absence of evidence is not evidence of absence: pitfalls of cre knock-ins in the c-Kit locus.Circ Res. 2014 Aug 1;115(4):415-8. doi: 10.1161/CIRCRESAHA.114.304676. Epub 2014 Jun 25. Circ Res. 2014. PMID: 24965482 No abstract available.
References
-
- Orlic D, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–705. - PubMed
-
- Balsam LB, et al. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668–673. - PubMed
-
- Murry CE, et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428:664–668. - PubMed
Additional references
-
- Nagy A, Rossant J. In: Gene Targeting:A Practical Approach. 2 edition. Joyner Alexandra L., editor. Oxford University Press, USA; 2000.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

