Genetic and genomic tools for the marine annelid Platynereis dumerilii
- PMID: 24807110
- PMCID: PMC4012478
- DOI: 10.1534/genetics.112.148254
Genetic and genomic tools for the marine annelid Platynereis dumerilii
Abstract
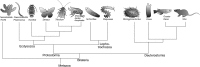
The bristle worm Platynereis dumerilii displays many interesting biological characteristics. These include its reproductive timing, which is synchronized to the moon phase, its regenerative capacity that is hormonally controlled, and a slow rate of evolution, which permits analyses of ancestral genes and cell types. As a marine annelid, Platynereis is also representative of the marine ecosystem, as well as one of the three large animal subphyla, the Lophotrochozoa. Here, we provide an overview of the molecular resources, functional techniques, and behavioral assays that have recently been established for the bristle worm. This combination of tools now places Platynereis in an excellent position to advance research at the frontiers of neurobiology, chronobiology, evo-devo, and marine biology.
Figures

References
-
- Ackermann C., 2003. Markierung der Zelllinien im Embryo von Platynereis. Ph.D. Thesis, University of Mainz, Mainz.
-
- Ackermann C., Dorresteijn A., Fischer A., 2005. Clonal domains in postlarval Platynereis dumerilii (Annelida: Polychaeta). J. Morphol. 266: 258–280. - PubMed
-
- Aguinaldo A. M., Turbeville J. M., Linford L. S., Rivera M. C., Garey J. R., et al. , 1997. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387: 489–493. - PubMed
-
- Arendt D., Technau U., Wittbrodt J., 2001. Evolution of the bilaterian larval foregut. Nature 409: 81–85. - PubMed
-
- Arendt D., Tessmar K., de Campos-Baptista M. I., Dorresteijn A., Wittbrodt J., 2002. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development 129: 1143–1154. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

