Activation of the pleiotropic drug resistance pathway can promote mitochondrial DNA retention by fusion-defective mitochondria in Saccharomyces cerevisiae
- PMID: 24807265
- PMCID: PMC4455774
- DOI: 10.1534/g3.114.010330
Activation of the pleiotropic drug resistance pathway can promote mitochondrial DNA retention by fusion-defective mitochondria in Saccharomyces cerevisiae
Abstract
Genetic and microscopic approaches using Saccharomyces cerevisiae have identified many proteins that play a role in mitochondrial dynamics, but it is possible that other proteins and pathways that play a role in mitochondrial division and fusion remain to be discovered. Mutants lacking mitochondrial fusion are characterized by rapid loss of mitochondrial DNA. We took advantage of a petite-negative mutant that is unable to survive mitochondrial DNA loss to select for mutations that allow cells with fusion-deficient mitochondria to maintain the mitochondrial genome on fermentable medium. Next-generation sequencing revealed that all identified suppressor mutations not associated with known mitochondrial division components were localized to PDR1 or PDR3, which encode transcription factors promoting drug resistance. Further studies revealed that at least one, if not all, of these suppressor mutations dominantly increases resistance to known substrates of the pleiotropic drug resistance pathway. Interestingly, hyperactivation of this pathway did not significantly affect mitochondrial shape, suggesting that mitochondrial division was not greatly affected. Our results reveal an intriguing genetic connection between pleiotropic drug resistance and mitochondrial dynamics.
Keywords: bulk segregant analysis; drug resistance; mitochondrial genome; mitochondrial shape; petite-negative.
Copyright © 2014 Mutlu et al.
Figures


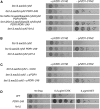
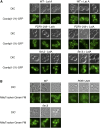
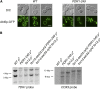
Similar articles
-
Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion.Biochem Biophys Res Commun. 2003 Aug 22;308(2):276-83. doi: 10.1016/s0006-291x(03)01348-2. Biochem Biophys Res Commun. 2003. PMID: 12901865
-
Mitochondrial anchorage and fusion contribute to mitochondrial inheritance and quality control in the budding yeast Saccharomyces cerevisiae.Mol Biol Cell. 2016 Mar 1;27(5):776-87. doi: 10.1091/mbc.E15-07-0455. Epub 2016 Jan 13. Mol Biol Cell. 2016. PMID: 26764088 Free PMC article.
-
Mitochondrial fusion increases the mitochondrial DNA copy number in budding yeast.Genes Cells. 2011 May;16(5):527-44. doi: 10.1111/j.1365-2443.2011.01504.x. Epub 2011 Apr 5. Genes Cells. 2011. PMID: 21463454
-
Mitochondrial dynamics in mammals.Curr Top Dev Biol. 2004;59:119-44. doi: 10.1016/S0070-2153(04)59005-1. Curr Top Dev Biol. 2004. PMID: 14975249 Review. No abstract available.
-
The power of yeast to model diseases of the powerhouse of the cell.Front Biosci (Landmark Ed). 2013 Jan 1;18(1):241-78. doi: 10.2741/4098. Front Biosci (Landmark Ed). 2013. PMID: 23276920 Free PMC article. Review.
Cited by
-
Evidence for Amino Acid Snorkeling from a High-Resolution, In Vivo Analysis of Fis1 Tail-Anchor Insertion at the Mitochondrial Outer Membrane.Genetics. 2017 Feb;205(2):691-705. doi: 10.1534/genetics.116.196428. Epub 2016 Dec 22. Genetics. 2017. PMID: 28007883 Free PMC article.
-
Identification and Characterization of Key Charged Residues in the Cofilin Protein Involved in Azole Susceptibility, Apoptosis, and Virulence of Aspergillus fumigatus.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e01659-17. doi: 10.1128/AAC.01659-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29483117 Free PMC article.
-
Bacterial tail anchors can target to the mitochondrial outer membrane.Biol Direct. 2017 Jul 24;12(1):16. doi: 10.1186/s13062-017-0187-0. Biol Direct. 2017. PMID: 28738827 Free PMC article.
-
Hybrid adaptation is hampered by Haldane's sieve.Nat Commun. 2024 Nov 28;15(1):10319. doi: 10.1038/s41467-024-54105-4. Nat Commun. 2024. PMID: 39609385 Free PMC article.
-
The contribution of Saccharomyces cerevisiae replicative age to the variations in the levels of Trx2p, Pdr5p, Can1p and Idh isoforms.Sci Rep. 2017 Oct 16;7(1):13220. doi: 10.1038/s41598-017-13576-w. Sci Rep. 2017. PMID: 29038504 Free PMC article.
References
-
- Adams A., Gottschling D., Kaiser C., Stearns T., 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
-
- Balzi E., Chen W., Ulaszewski S., Capieaux E., Goffeau A., 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262: 16871–16879. - PubMed
-
- Balzi E., Wang M., Leterme S., Van Dyck L., Goffeau A., 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269: 2206–2214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
