Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs
- PMID: 24807715
- PMCID: PMC4097787
- DOI: 10.1128/JVI.00985-14
Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs
Abstract
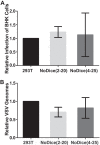
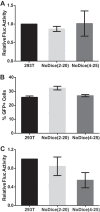
The issue of whether viruses are subject to restriction by endogenous microRNAs (miRNAs) and/or by virus-induced small interfering RNAs (siRNAs) in infected human somatic cells has been controversial. Here, we address this question in two ways. First, using deep sequencing, we demonstrate that infection of human cells by the RNA virus dengue virus (DENV) or West Nile virus (WNV) does not result in the production of any virus-derived siRNAs or viral miRNAs. Second, to more globally assess the potential of small regulatory RNAs to inhibit virus replication, we used gene editing to derive human cell lines that lack a functional Dicer enzyme and that therefore are unable to produce miRNAs or siRNAs. Infection of these cells with a wide range of viruses, including DENV, WNV, yellow fever virus, Sindbis virus, Venezuelan equine encephalitis virus, measles virus, influenza A virus, reovirus, vesicular stomatitis virus, human immunodeficiency virus type 1, or herpes simplex virus 1 (HSV-1), failed to reveal any enhancement in the replication of any of these viruses, although HSV-1, which encodes at least eight Dicer-dependent viral miRNAs, did replicate somewhat more slowly in the absence of Dicer. We conclude that most, and perhaps all, human viruses have evolved to be resistant to inhibition by endogenous human miRNAs during productive replication and that dependence on a cellular miRNA, as seen with hepatitis C virus, is rare. How viruses have evolved to avoid inhibition by endogenous cellular miRNAs, which are generally highly conserved during metazoan evolution, remains to be determined. Importance: Eukaryotic cells express a wide range of small regulatory RNAs, including miRNAs, that have the potential to inhibit the expression of mRNAs that show sequence complementarity. Indeed, previous work has suggested that endogenous miRNAs have the potential to inhibit viral gene expression and replication. Here, we demonstrate that the replication of a wide range of pathogenic viruses is not enhanced in human cells engineered to be unable to produce miRNAs, indicating that viruses have evolved to be resistant to inhibition by miRNAs. This result is important, as it implies that manipulation of miRNA levels is not likely to prove useful in inhibiting virus replication. It also focuses attention on the question of how viruses have evolved to resist inhibition by miRNAs and whether virus mutants that have lost this resistance might prove useful, for example, in the development of attenuated virus vaccines.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007392/AI/NIAID NIH HHS/United States
- R01-DA0300086/DA/NIDA NIH HHS/United States
- T32 GM007184/GM/NIGMS NIH HHS/United States
- T32 CA009111/CA/NCI NIH HHS/United States
- T32-AI007392/AI/NIAID NIH HHS/United States
- R01-NS038932/NS/NINDS NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- R01 NS038932/NS/NINDS NIH HHS/United States
- U54-AI057157/AI/NIAID NIH HHS/United States
- U54 AI057157/AI/NIAID NIH HHS/United States
- R01-AI083333/AI/NIAID NIH HHS/United States
- R01-AI0973376/AI/NIAID NIH HHS/United States
- T32-AI007247/AI/NIAID NIH HHS/United States
- R01 AI089526/AI/NIAID NIH HHS/United States
- R01-AI089526/AI/NIAID NIH HHS/United States
- T32-CA009111/CA/NCI NIH HHS/United States
- T32 AI007247/AI/NIAID NIH HHS/United States
- R01 DA030086/DA/NIDA NIH HHS/United States
- R01 AI083333/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

