The varicella-zoster virus portal protein is essential for cleavage and packaging of viral DNA
- PMID: 24807720
- PMCID: PMC4097804
- DOI: 10.1128/JVI.00376-14
The varicella-zoster virus portal protein is essential for cleavage and packaging of viral DNA
Abstract
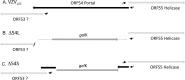
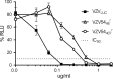
The varicella-zoster virus (VZV) open reading frame 54 (ORF54) gene encodes an 87-kDa monomer that oligomerizes to form the VZV portal protein, pORF54. pORF54 was hypothesized to perform a function similar to that of a previously described herpes simplex virus 1 (HSV-1) homolog, pUL6. pUL6 and the associated viral terminase are required for processing of concatemeric viral DNA and packaging of individual viral genomes into preformed capsids. In this report, we describe two VZV bacterial artificial chromosome (BAC) constructs with ORF54 gene deletions, Δ54L (full ORF deletion) and Δ54S (partial internal deletion). The full deletion of ORF54 likely disrupted essential adjacent genes (ORF53 and ORF55) and therefore could not be complemented on an ORF54-expressing cell line (ARPE54). In contrast, Δ54S was successfully propagated in ARPE54 cells but failed to replicate in parental, noncomplementing ARPE19 cells. Transmission electron microscopy confirmed the presence of only empty VZV capsids in Δ54S-infected ARPE19 cell nuclei. Similar to the HSV-1 genome, the VZV genome is composed of a unique long region (UL) and a unique short region (US) flanked by inverted repeats. DNA from cells infected with parental VZV (VZVLUC strain) contained the predicted UL and US termini, whereas cells infected with Δ54S contained neither. This result demonstrates that Δ54S is not able to process and package viral DNA, thus making pORF54 an excellent chemotherapeutic target. In addition, the utility of BAC constructs Δ54L and Δ54S as tools for the isolation of site-directed ORF54 mutants was demonstrated by recombineering single-nucleotide changes within ORF54 that conferred resistance to VZV-specific portal protein inhibitors. Importance: Antivirals with novel mechanisms of action would provide additional therapeutic options to treat human herpesvirus infections. Proteins involved in the herpesviral DNA encapsidation process have become promising antiviral targets. Previously, we described a series of N-α-methylbenzyl-N'-aryl thiourea analogs that target the VZV portal protein (pORF54) and prevent viral replication in vitro. To better understand the mechanism of action of these compounds, it is important to define the structural and functional characteristics of the VZV portal protein. In contrast to HSV, no VZV mutants have been described for any of the seven essential DNA encapsidation genes. The VZV ORF54 deletion mutant described in this study represents the first VZV encapsidation mutant reported to date. We demonstrate that the deletion mutant can serve as a platform for the isolation of portal mutants via recombineering and provide a strategy for more in-depth studies of VZV portal structure and function.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures







Similar articles
-
Mutagenesis and functional analysis of the varicella-zoster virus portal protein.J Virol. 2024 Apr 16;98(4):e0060323. doi: 10.1128/jvi.00603-23. Epub 2024 Mar 22. J Virol. 2024. PMID: 38517165 Free PMC article.
-
Intermolecular Complementation between Two Varicella-Zoster Virus pORF30 Terminase Domains Essential for DNA Encapsidation.J Virol. 2015 Oct;89(19):10010-22. doi: 10.1128/JVI.01313-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202238 Free PMC article.
-
Role of the Herpes Simplex Virus CVSC Proteins at the Capsid Portal Vertex.J Virol. 2020 Nov 23;94(24):e01534-20. doi: 10.1128/JVI.01534-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967953 Free PMC article.
-
Mutagenesis of the varicella-zoster virus genome: lessons learned.Arch Virol Suppl. 2001;(17):91-7. doi: 10.1007/978-3-7091-6259-0_10. Arch Virol Suppl. 2001. PMID: 11339555 Review.
-
Human herpesvirus portal proteins: Structure, function, and antiviral prospects.Rev Med Virol. 2018 May;28(3):e1972. doi: 10.1002/rmv.1972. Epub 2018 Mar 24. Rev Med Virol. 2018. PMID: 29573302 Review.
Cited by
-
Cryo-EM structure of the varicella-zoster virus A-capsid.Nat Commun. 2020 Sep 22;11(1):4795. doi: 10.1038/s41467-020-18537-y. Nat Commun. 2020. PMID: 32963252 Free PMC article.
-
Mutagenesis and functional analysis of the varicella-zoster virus portal protein.J Virol. 2024 Apr 16;98(4):e0060323. doi: 10.1128/jvi.00603-23. Epub 2024 Mar 22. J Virol. 2024. PMID: 38517165 Free PMC article.
-
Intermolecular Complementation between Two Varicella-Zoster Virus pORF30 Terminase Domains Essential for DNA Encapsidation.J Virol. 2015 Oct;89(19):10010-22. doi: 10.1128/JVI.01313-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202238 Free PMC article.
-
Effects of Shield1 on the viral replication of varicella‑zoster virus containing FKBP‑tagged ORF4 and 48.Mol Med Rep. 2018 Jan;17(1):763-770. doi: 10.3892/mmr.2017.7986. Epub 2017 Nov 6. Mol Med Rep. 2018. PMID: 29115621 Free PMC article.
-
Congenital and Perinatal Varicella Infections.Newborn (Clarksville). 2022 Jul-Sep;1(3):278-286. doi: 10.5005/jp-journals-11002-0040. Epub 2022 Oct 7. Newborn (Clarksville). 2022. PMID: 36540194 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

