CYTOKININ OXIDASE/DEHYDROGENASE4 Integrates Cytokinin and Auxin Signaling to Control Rice Crown Root Formation
- PMID: 24808099
- PMCID: PMC4081320
- DOI: 10.1104/pp.114.238584
CYTOKININ OXIDASE/DEHYDROGENASE4 Integrates Cytokinin and Auxin Signaling to Control Rice Crown Root Formation
Abstract
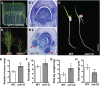
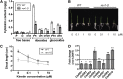
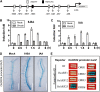
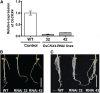
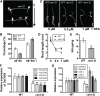
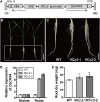
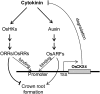
Crown roots constitute the majority of the rice (Oryza sativa) root system and play an important role in rice growth and development. However, the molecular mechanism of crown root formation in rice is not well understood. Here, we characterized a rice dominant mutant, root enhancer1 (ren1-D), which was observed to exhibit a more robust root system, increased crown root number, and reduced plant height. Molecular and genetic analyses revealed that these phenotypes are caused by the activation of a cytokinin oxidase/dehydrogenase (CKX) family gene, OsCKX4. Subcellular localization demonstrated that OsCKX4 is a cytosolic isoform of CKX. OsCKX4 is predominantly expressed in leaf blades and roots. It is the dominant CKX, preferentially expressed in the shoot base where crown root primordia are produced, underlining its role in root initiation. OsCKX4 is induced by exogenous auxin and cytokinin in the roots. Furthermore, one-hybrid assays revealed that OsCKX4 is a direct binding target of both the auxin response factor OsARF25 and the cytokinin response regulators OsRR2 and OsRR3. Overexpression and RNA interference of OsCKX4 confirmed that OsCKX4 plays a positive role in crown root formation. Moreover, expression analysis revealed a significant alteration in the expression of auxin-related genes in the ren1-D mutants, indicating that the OsCKX4 mediates crown root development by integrating the interaction between cytokinin and auxin. Transgenic plants harboring OsCKX4 under the control of the root-specific promoter RCc3 displayed enhanced root development without affecting their shoot parts, suggesting that this strategy could be a powerful tool in rice root engineering.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 - PubMed
-
- Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, Wang J, Yang Y, Zhu M. (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol 196: 149–161 - PubMed
-
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P. (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci 15: 219–226 - PubMed
-
- de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12: 474–481 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

