Insight into the role of substrate-binding residues in conferring substrate specificity for the multifunctional polysaccharide lyase Smlt1473
- PMID: 24808176
- PMCID: PMC4140301
- DOI: 10.1074/jbc.M114.571299
Insight into the role of substrate-binding residues in conferring substrate specificity for the multifunctional polysaccharide lyase Smlt1473
Abstract
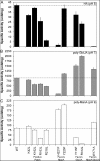
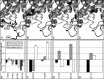
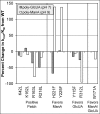
Anionic polysaccharides are of growing interest in the biotechnology industry due to their potential pharmaceutical applications in drug delivery and wound treatment. Chemical composition and polymer length strongly influence the physical and biological properties of the polysaccharide and thus its potential industrial and medical applications. One promising approach to determining monomer composition and controlling the degree of polymerization involves the use of polysaccharide lyases, which catalyze the depolymerization of anionic polysaccharides via a β-elimination mechanism. Utilization of these enzymes for the production of custom-made oligosaccharides requires a high degree of control over substrate specificity. Previously, we characterized a polysaccharide lyase (Smlt1473) from Stenotrophomonas maltophilia k279a, which exhibited significant activity against hyaluronan (HA), poly-β-d-glucuronic acid (poly-GlcUA), and poly-β-d-mannuronic acid (poly-ManA) in a pH-regulated manner. Here, we utilize a sequence structure guided approach based on a homology model of Smlt1473 to identify nine putative substrate-binding residues and examine their effect on substrate specificity via site-directed mutagenesis. Interestingly, single point mutations H221F and R312L resulted in increased activity and specificity toward poly-ManA and poly-GlcUA, respectively. Furthermore, a W171A mutant nearly eliminated HA activity, while increasing poly-ManA and poly-GlcUA activity by at least 35%. The effect of these mutations was analyzed by comparison with the high resolution structure of Sphingomonas sp. A1-III alginate lyase in complex with poly-ManA tetrasaccharide and by taking into account the structural differences between HA, poly-GlcUA, and poly-ManA. Overall, our results demonstrate that even minor changes in active site architecture have a significant effect on the substrate specificity of Smlt1473, whose structural plasticity could be applied to the design of highly active and specific polysaccharide lyases.
Keywords: Carbohydrate Processing; Enzyme Mechanism; Enzyme Mutation; Lyase; Polysaccharide; Protein Design; Substrate Specificity.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

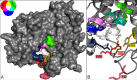

References
-
- Gacesa P. (1987) Alginate-modifying enzymes: A proposed unified mechanism of action for the lyases and epimerases. FEBS Lett. 212, 199–202
-
- Garron M. L., Cygler M. (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20, 1547–1573 - PubMed
-
- Yoon H. J., Hashimoto W., Miyake O., Murata K., Mikami B. (2001) Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution. J. Mol. Biol. 307, 9–16 - PubMed
-
- Maruyama Y., Hashimoto W., Mikami B., Murata K. (2005) Crystal structure of Bacillus sp. GL1 xanthan lyase complexed with a substrate: insights into the enzyme reaction mechanism. J. Mol. Biol. 350, 974–986 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

