Genome-wide (over)view on the actions of vitamin D
- PMID: 24808867
- PMCID: PMC4010781
- DOI: 10.3389/fphys.2014.00167
Genome-wide (over)view on the actions of vitamin D
Abstract
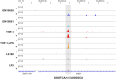
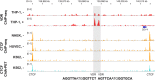
For a global understanding of the physiological impact of the nuclear hormone 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) the analysis of the genome-wide locations of its high affinity receptor, the transcription factor vitamin D receptor (VDR), is essential. Chromatin immunoprecipitation sequencing (ChIP-seq) in GM10855 and GM10861 lymphoblastoid cells, undifferentiated and lipopolysaccharide-differentiated THP-1 monocytes, LS180 colorectal cancer cells and LX2 hepatic stellate cells revealed between 1000 and 13,000 VDR-specific genomic binding sites. The harmonized analysis of these ChIP-seq datasets indicates that the mechanistic basis for the action of the VDR is independent of the cell type. Formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq) data highlight accessible chromatin regions, which are under control of 1,25(OH)2D3. In addition, public data, such as from the ENCODE project, allow to relate the genome-wide actions of VDR and 1,25(OH)2D3 to those of other proteins within the nucleus. For example, locations of the insulator protein CTCF suggest a segregation of the human genome into chromatin domains, of which more than 1000 contain at least one VDR binding site. The integration of all these genome-wide data facilitates the identification of the most important VDR binding sites and associated primary 1,25(OH)2D3 target genes. Expression changes of these key genes can serve as biomarkers for the actions of vitamin D3 and its metabolites in different tissues and cell types of human individuals. Analysis of primary tissues obtained from vitamin D3 intervention studies using such markers indicated a large inter-individual variation for the efficiency of vitamin D3 supplementation. In conclusion, a genome-wide (over)view on the genomic locations of VDR provides a broader basis for addressing vitamin D's role in health and disease.
Keywords: chromatin; epigenomics; gene regulation; genomics; vitamin D; vitamin D receptor.
Figures

References
-
- Carlberg C., Bendik I., Wyss A., Meier E., Sturzenbecker L. J., Grippo J. F., Hunziker W. (1993). Two nuclear signalling pathways for vitamin D. Nature 361, 657–660 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

