Redefining the modular organization of the core Mediator complex
- PMID: 24810298
- PMCID: PMC4085763
- DOI: 10.1038/cr.2014.64
Redefining the modular organization of the core Mediator complex
Abstract
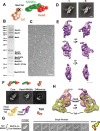
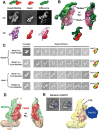
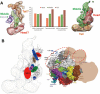
The Mediator complex plays an essential role in the regulation of eukaryotic transcription. The Saccharomyces cerevisiae core Mediator comprises 21 subunits, which are organized into Head, Middle and Tail modules. Previously, the Head module was assigned to a distinct dense domain at the base, and the Middle and Tail modules were identified to form a tight structure above the Head module, which apparently contradicted findings from many biochemical and functional studies. Here, we compared the structures of the core Mediator and its subcomplexes, especially the first 3D structure of the Head + Middle modules, which permitted an unambiguous assignment of the three modules. Furthermore, nanogold labeling pinpointing four Mediator subunits from different modules conclusively validated the modular assignment, in which the Head and Middle modules fold back on one another and form the upper portion of the core Mediator, while the Tail module forms a distinct dense domain at the base. The new modular model of the core Mediator has reconciled the previous inconsistencies between the structurally and functionally defined Mediator modules. Collectively, these analyses completely redefine the modular organization of the core Mediator, which allow us to integrate the structural and functional information into a coherent mechanism for the Mediator's modularity and regulation in transcription initiation.
Figures

Comment in
-
Mediator redefines itself.Cell Res. 2014 Jul;24(7):775-6. doi: 10.1038/cr.2014.76. Epub 2014 Jun 10. Cell Res. 2014. PMID: 24913192 Free PMC article.
References
-
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. - PubMed
-
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. - PubMed
-
- Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. - PubMed
-
- Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

