The DAN family: modulators of TGF-β signaling and beyond
- PMID: 24810382
- PMCID: PMC4116650
- DOI: 10.1002/pro.2485
The DAN family: modulators of TGF-β signaling and beyond
Abstract
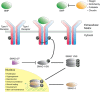
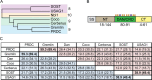
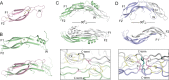
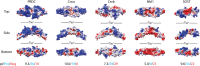
Extracellular binding proteins or antagonists are important factors that modulate ligands in the transforming growth factor (TGF-β) family. While the interplay between antagonists and ligands are essential for developmental and normal cellular processes, their imbalance can lead to the pathology of several disease states. In particular, recent studies have implicated members of the differential screening-selected gene in neuroblastoma (DAN) family in disease such as renal fibrosis, pulmonary arterial hypertension, and reactivation of metastatic cancer stem cells. DAN family members are known to inhibit the bone morphogenetic proteins (BMP) of the TGF-β family. However, unlike other TGF-β antagonist families, DAN family members have roles beyond ligand inhibition and can modulate Wnt and vascular endothelial growth factor (VEGF) signaling pathways. This review describes recent structural and functional advances that have expanded our understanding of DAN family proteins with regards to BMP inhibition and also highlights their emerging roles in the modulation of Wnt and VEGF signaling pathways.
Keywords: BMP; Keywords DAN; TGF-β; VEGF; Wnt; extracellular antagonists.
© 2014 The Protein Society.
Figures



References
-
- Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6:432–438. - PubMed
-
- Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23:609–620. - PubMed
-
- Rider CCC, Mulloy BB. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J. 2010;429:1–12. - PubMed
-
- Walsh DW, Godson C, Brazil DP, Martin F. Extracellular BMP-antagonist regulation in development and disease: tied up in knots. Trends Cell Biol. 2010;20:244–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

