e-RNA: a collection of web servers for comparative RNA structure prediction and visualisation
- PMID: 24810851
- PMCID: PMC4086097
- DOI: 10.1093/nar/gku292
e-RNA: a collection of web servers for comparative RNA structure prediction and visualisation
Abstract
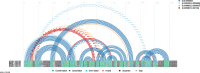
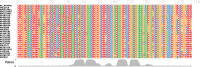
e-RNA offers a free and open-access collection of five published RNA sequence analysis tools, each solving specific problems not readily addressed by other available tools. Given multiple sequence alignments, Transat detects all conserved helices, including those expected in a final structure, but also transient, alternative and pseudo-knotted helices. RNA-Decoder uses unique evolutionary models to detect conserved RNA secondary structure in alignments which may be partly protein-coding. SimulFold simultaneously co-estimates the potentially pseudo-knotted conserved structure, alignment and phylogenetic tree for a set of homologous input sequences. CoFold predicts the minimum-free energy structure for an input sequence while taking the effects of co-transcriptional folding into account, thereby greatly improving the prediction accuracy for long sequences. R-chie is a program to visualise RNA secondary structures as arc diagrams, allowing for easy comparison and analysis of conserved base-pairs and quantitative features. The web site server dispatches user jobs to a cluster, where up to 100 jobs can be processed in parallel. Upon job completion, users can retrieve their results via a bookmarked or emailed link. e-RNA is located at http://www.e-rna.org.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
e-RNA: a collection of web-servers for the prediction and visualisation of RNA secondary structure and their functional features.Nucleic Acids Res. 2023 Jul 5;51(W1):W160-W167. doi: 10.1093/nar/gkad296. Nucleic Acids Res. 2023. PMID: 37099369 Free PMC article.
-
R-CHIE: a web server and R package for visualizing RNA secondary structures.Nucleic Acids Res. 2012 Jul;40(12):e95. doi: 10.1093/nar/gks241. Epub 2012 Mar 19. Nucleic Acids Res. 2012. PMID: 22434875 Free PMC article.
-
TRANSAT-- method for detecting the conserved helices of functional RNA structures, including transient, pseudo-knotted and alternative structures.PLoS Comput Biol. 2010 Jun 24;6(6):e1000823. doi: 10.1371/journal.pcbi.1000823. PLoS Comput Biol. 2010. PMID: 20589081 Free PMC article.
-
RNA-TVcurve: a Web server for RNA secondary structure comparison based on a multi-scale similarity of its triple vector curve representation.BMC Bioinformatics. 2017 Jan 21;18(1):51. doi: 10.1186/s12859-017-1481-7. BMC Bioinformatics. 2017. PMID: 28109252 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
-
The International Virus Bioinformatics Meeting 2020.Viruses. 2020 Dec 6;12(12):1398. doi: 10.3390/v12121398. Viruses. 2020. PMID: 33291220 Free PMC article.
-
The dynamic proteome of influenza A virus infection identifies M segment splicing as a host range determinant.Nat Commun. 2019 Dec 4;10(1):5518. doi: 10.1038/s41467-019-13520-8. Nat Commun. 2019. PMID: 31797923 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

