Mutations in the X-linked intellectual disability gene, zDHHC9, alter autopalmitoylation activity by distinct mechanisms
- PMID: 24811172
- PMCID: PMC4140262
- DOI: 10.1074/jbc.M114.567420
Mutations in the X-linked intellectual disability gene, zDHHC9, alter autopalmitoylation activity by distinct mechanisms
Abstract
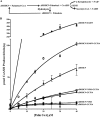
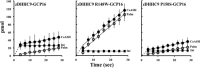
Early onset intellectual disabilities result in significant societal and economic costs and affect 1-3% of the population. The underlying genetic determinants are beginning to emerge and are interpreted in the context of years of work characterizing postsynaptic receptor and signaling functions of learning and memory. DNA sequence analysis of intellectual disability patients has revealed greater than 80 loci on the X-chromosome that are potentially linked to disease. One of the loci is zDHHC9, a gene encoding a Ras protein acyltransferase. Protein palmitoylation is a reversible modification that controls the subcellular localization and distribution of membrane receptors, scaffolds, and signaling proteins required for neuronal plasticity. Palmitoylation occurs in two steps. In the first step, autopalmitoylation, an enzyme-palmitoyl intermediate is formed. During the second step, the palmitoyl moiety is transferred to a protein substrate, or if no substrate is available, hydrolysis of the thioester linkage produces the enzyme and free palmitate. In this study, we demonstrate that two naturally occurring variants of zDHHC9, encoding R148W and P150S, affect the autopalmitoylation step of the reaction by lowering the steady state amount of the palmitoyl-zDHHC9 intermediate.
Keywords: Enzyme Mechanism; Post-translational Modification (PTM); Protein Acylation; Protein Palmitoylation; Ras Protein; X-linked Intellectual Disability; zDHHC Proteins.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Rocks O., Gerauer M., Vartak N., Koch S., Huang Z. P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P. I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 - PubMed
-
- Fukata Y., Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 - PubMed
-
- Zhang W., Trible R. P., Samelson L. E. (1998) LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity 9, 239–246 - PubMed
-
- Joseph M., Nagaraj R. (1995) Conformations of peptides corresponding to fatty acylation sites in proteins. A circular dichroism study. J. Biol. Chem. 270, 19439–19445 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

