FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma
- PMID: 24811343
- PMCID: PMC5827933
- DOI: 10.1038/leu.2014.126
FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma
Abstract
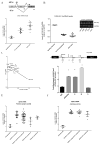
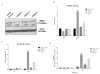
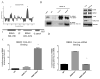
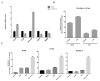
Impaired Fas-mediated apoptosis is associated with poor clinical outcomes and cancer chemoresistance. Soluble Fas receptor (sFas), produced by skipping of exon 6, inhibits apoptosis by sequestering Fas ligand. Serum sFas is associated with poor prognosis of non-Hodgkin's lymphomas. We found that the alternative splicing of Fas in lymphomas is tightly regulated by a long-noncoding RNA corresponding to an antisense transcript of Fas (FAS-AS1). Levels of FAS-AS1 correlate inversely with production of sFas, and FAS-AS1 binding to the RBM5 inhibits RBM5-mediated exon 6 skipping. EZH2, often mutated or overexpressed in lymphomas, hyper-methylates the FAS-AS1 promoter and represses the FAS-AS1 expression. EZH2-mediated repression of FAS-AS1 promoter can be released by DZNeP (3-Deazaneplanocin A) or overcome by ectopic expression of FAS-AS1, both of which increase levels of FAS-AS1 and correspondingly decrease expression of sFas. Treatment with Bruton's tyrosine kinase inhibitor or EZH2 knockdown decreases the levels of EZH2, RBM5 and sFas, thereby enhancing Fas-mediated apoptosis. This is the first report showing functional regulation of Fas repression by its antisense RNA. Our results reveal new therapeutic targets in lymphomas and provide a rationale for the use of EZH2 inhibitors or ibrutinib in combination with chemotherapeutic agents that recruit Fas for effective cell killing.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Nagata S. Fas and Fas ligand: a death factor and its receptor. Advances in Immunology. 1994;57:129–144. - PubMed
-
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995 Mar 10;267(5203):1456–1462. - PubMed
-
- Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000 Mar;21(3):485–495. - PubMed
-
- Rudin CM, Thompson CB. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annual Review of Medicine. 1997;48:267–281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

