Cystoviral polymerase complex protein P7 uses its acidic C-terminal tail to regulate the RNA-directed RNA polymerase P2
- PMID: 24813120
- PMCID: PMC4090703
- DOI: 10.1016/j.jmb.2014.04.028
Cystoviral polymerase complex protein P7 uses its acidic C-terminal tail to regulate the RNA-directed RNA polymerase P2
Abstract
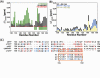
In bacteriophages of the cystovirus family, the polymerase complex (PX) encodes a 75-kDa RNA-directed RNA polymerase (P2) that transcribes the double-stranded RNA genome. Also a constituent of the PX is the essential protein P7 that, in addition to accelerating PX assembly and facilitating genome packaging, plays a regulatory role in transcription. Deletion of P7 from the PX leads to aberrant plus-strand synthesis suggesting its influence on the transcriptase activity of P2. Here, using solution NMR techniques and the P2 and P7 proteins from cystovirus ϕ12, we demonstrate their largely electrostatic interaction in vitro. Chemical shift perturbations on P7 in the presence of P2 suggest that this interaction involves the dynamic C-terminal tail of P7, more specifically an acidic cluster therein. Patterns of chemical shift changes induced on P2 by the P7 C-terminus resemble those seen in the presence of single-stranded RNA suggesting similarities in binding. This association between P2 and P7 reduces the affinity of the former toward template RNA and results in its decreased activity both in de novo RNA synthesis and in extending a short primer. Given the presence of C-terminal acidic tracts on all cystoviral P7 proteins, the electrostatic nature of the P2/P7 interaction is likely conserved within the family and could constitute a mechanism through which P7 regulates transcription in cystoviruses.
Keywords: NMR; RNA-directed RNA polymerase; cystovirus; polymerase complex; transcription.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


References
-
- Mindich L, Bamford DH. Lipid-containing bacteriophages. In: Calendar R, editor. The Bacteriophages. Plenum Publishing Corporation; New York: 1988. pp. 475–519.
-
- Hoogstraten D, Qiao X, Sun Y, Hu A, Onodera S, Mindich L. Characterization of ϕ8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to ϕ6. Virology. 2000;272:218–24. - PubMed
-
- Qiao X, Qiao J, Onodera S, Mindich L. Characterization of ϕ13, a bacteriophage related to ϕ6 and containing three dsRNA genomic segments. Virology. 2000;275:218–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

