Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells
- PMID: 24813441
- PMCID: PMC4066773
- DOI: 10.1093/nar/gku346
Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells
Abstract
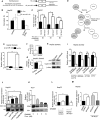
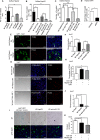
miRNAs are 20-22 nt long post-transcriptional regulators in metazoan cells that repress protein expression from their target mRNAs. These tiny regulatory RNAs follow tissue and cell-type specific expression pattern, aberrations of which are associated with various diseases. miR-122 is a liver-specific anti-proliferative miRNA that, we found, can be transferred via exosomes between human hepatoma cells, Huh7 and HepG2, grown in co-culture. Exosomal miR-122, expressed and released by Huh7 cells and taken by miR-122 deficient HepG2 cells, was found to be effective in repression of target mRNAs and to reduce growth and proliferation of recipient HepG2 cells. Interestingly, in a reciprocal process, HepG2 secretes Insulin-like Growth Factor 1 (IGF1) that decreases miR-122 expression in Huh7 cells. Our observations suggest existence of a reciprocal interaction between two different hepatic cells with distinct miR-122 expression profiles. This interaction is mediated via intercellular exosome-mediated miR-122 transfer and countered by a reciprocal IGF1-dependent anti-miR-122 signal. According to our data, human hepatoma cells use IGF1 to prevent intercellular exosomal transfer of miR-122 to ensure its own proliferation by preventing expression of growth retarding miR-122 in neighbouring cells.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. - PubMed
-
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

