Phylogeny of a relapsing fever Borrelia species transmitted by the hard tick Ixodes scapularis
- PMID: 24813576
- PMCID: PMC4182126
- DOI: 10.1016/j.meegid.2014.04.022
Phylogeny of a relapsing fever Borrelia species transmitted by the hard tick Ixodes scapularis
Abstract
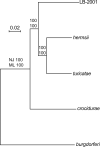
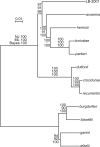
The discovery of Borrelia species that were related to the agents of relapsing fever but were transmitted by hard ticks rather than soft ticks challenged previous taxonomies based largely on microbe-host specificities and geographic considerations. One of these newly-identified organisms is the Borrelia miyamotoi sensu lato strain LB-2001 from North America and transmitted by Ixodes scapularis. This or related strains have been identified as the cause of human disease, but comparatively little is known about their biology or genetics. Using recently acquired chromosome sequence of LB-2001 together with database sequences and additional sequences determined here, I carried out comparisons of the several species of Borrelia, including those in the two major clades: the relapsing fever group of species and the Lyme disease group of species. Phylogenetic inference at the species level was based on four data sets: whole chromosomes of ∼1Mb each, and concatenated sequences of 19 ribosomal protein genes, 3 conserved nucleic acid enzymes (rpoC, recC, and dnaE), and 4 contiguous genes for nucleotide salvage on a large plasmid. Analyses using neighbor-joining, maximum likelihood, and Bayesian methods were largely concordant for each of the trees. They showed that LB-2001 and related hard tick-associated organisms, like Borrelia lonestari, are deeply positioned within the RF group of species and that these organisms did not, as some earlier estimations had suggested, constitute a paraphyletic group. The analyses also provided further evidence that major changes in host ranges and life cycles, such as hard to soft ticks or vice versa, may not correlate well with overall sequence differences. The genetic differences between LB-2001 and B. miyamotoi sensu stricto justify provisional use of the "sensu lato" designation for LB-2001.
Keywords: Genomics; Lyme disease; Spirochetes; Tick-borne disease.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures





References
-
- Barbour AG. Borrelia: a diverse and ubiquitous genus of tick-borne pathogens. In: Scheld WM, Craig WA, Hughes JM, editors. Emerging Infections 5. ASM Press; Washington, D.C.: 2001. pp. 153–174.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

