Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears
- PMID: 24813606
- PMCID: PMC4089990
- DOI: 10.1016/j.cell.2014.03.054
Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears
Abstract
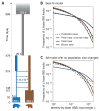
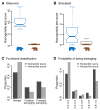
Polar bears are uniquely adapted to life in the High Arctic and have undergone drastic physiological changes in response to Arctic climates and a hyper-lipid diet of primarily marine mammal prey. We analyzed 89 complete genomes of polar bear and brown bear using population genomic modeling and show that the species diverged only 479-343 thousand years BP. We find that genes on the polar bear lineage have been under stronger positive selection than in brown bears; nine of the top 16 genes under strong positive selection are associated with cardiomyopathy and vascular disease, implying important reorganization of the cardiovascular system. One of the genes showing the strongest evidence of selection, APOB, encodes the primary lipoprotein component of low-density lipoprotein (LDL); functional mutations in APOB may explain how polar bears are able to cope with life-long elevated LDL levels that are associated with high risk of heart disease in humans.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Genome evolution: Evolutionary insights from comparative bear genomics.Nat Rev Genet. 2014 Jul;15(7):442. doi: 10.1038/nrg3757. Epub 2014 May 20. Nat Rev Genet. 2014. PMID: 24840550 No abstract available.
References
-
- Atkinson SN, Ramsay MA. The effects of prolonged fasting of the body composition and reproductive success of female polar bears (Ursus maritimus) Funct Ecol. 1995;9:559–567.
-
- Atkinson SN, Nelson RA, Ramsay MA. Changes in the body composition of fasting polar bears (Ursus maritimus): the effect of relative fatness on protein conservation. Physiol Zool. 1996;69:304–316.
-
- Benn M. Apolipoprotein B levels, APOB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis. 2009;206:17–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

