Programming and inheritance of parental DNA methylomes in mammals
- PMID: 24813617
- PMCID: PMC4096154
- DOI: 10.1016/j.cell.2014.04.017
Programming and inheritance of parental DNA methylomes in mammals
Erratum in
- Cell. 2014 Jun 19;157(7):1735
Abstract
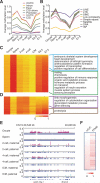
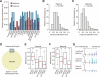
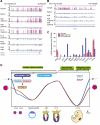
The reprogramming of parental methylomes is essential for embryonic development. In mammals, paternal 5-methylcytosines (5mCs) have been proposed to be actively converted to oxidized bases. These paternal oxidized bases and maternal 5mCs are believed to be passively diluted by cell divisions. By generating single-base resolution, allele-specific DNA methylomes from mouse gametes, early embryos, and primordial germ cell (PGC), as well as single-base-resolution maps of oxidized cytosine bases for early embryos, we report the existence of 5hmC and 5fC in both maternal and paternal genomes and find that 5mC or its oxidized derivatives, at the majority of demethylated CpGs, are converted to unmodified cytosines independent of passive dilution from gametes to four-cell embryos. Therefore, we conclude that paternal methylome and at least a significant proportion of maternal methylome go through active demethylation during embryonic development. Additionally, all the known imprinting control regions (ICRs) were classified into germ-line or somatic ICRs.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Collado-Fernandez E, Picton HM, Dumollard R. Metabolism throughout follicle and oocyte development in mammals. The International journal of developmental biology. 2012;56:799–808. - PubMed
-
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. - PubMed
-
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

