Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment
- PMID: 24814466
- PMCID: PMC4096375
- DOI: 10.1534/genetics.114.163147
Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment
Abstract
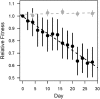
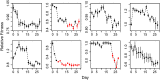
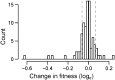
Our understanding of the evolutionary consequences of mutation relies heavily on estimates of the rate and fitness effect of spontaneous mutations generated by mutation accumulation (MA) experiments. We performed a classic MA experiment in which frequent sampling of MA lines was combined with whole genome resequencing to develop a high-resolution picture of the effect of spontaneous mutations in a hypermutator (ΔmutS) strain of the bacterium Pseudomonas aeruginosa. After ∼644 generations of mutation accumulation, MA lines had accumulated an average of 118 mutations, and we found that average fitness across all lines decayed linearly over time. Detailed analyses of the dynamics of fitness change in individual lines revealed that a large fraction of the total decay in fitness (42.3%) was attributable to the fixation of rare, highly deleterious mutations (comprising only 0.5% of fixed mutations). Furthermore, we found that at least 0.64% of mutations were beneficial and probably fixed due to positive selection. The majority of mutations that fixed (82.4%) were base substitutions and we failed to find any signatures of selection on nonsynonymous or intergenic mutations. Short indels made up a much smaller fraction of the mutations that were fixed (17.4%), but we found evidence of strong selection against indels that caused frameshift mutations in coding regions. These results help to quantify the amount of natural selection present in microbial MA experiments and demonstrate that changes in fitness are strongly influenced by rare mutations of large effect.
Keywords: Pseudomonas aeruginosa; experimental evolution; hypermutator; spontaneous mutation; whole genome resequencing.
Copyright © 2014 by the Genetics Society of America.
Figures
Similar articles
-
Mutational Landscape of Spontaneous Base Substitutions and Small Indels in Experimental Caenorhabditis elegans Populations of Differing Size.Genetics. 2019 Jul;212(3):837-854. doi: 10.1534/genetics.119.302054. Epub 2019 May 20. Genetics. 2019. PMID: 31110155 Free PMC article.
-
The Fitness Effects of Spontaneous Mutations Nearly Unseen by Selection in a Bacterium with Multiple Chromosomes.Genetics. 2016 Nov;204(3):1225-1238. doi: 10.1534/genetics.116.193060. Epub 2016 Sep 26. Genetics. 2016. PMID: 27672096 Free PMC article.
-
Sequence context of indel mutations and their effect on protein evolution in a bacterial endosymbiont.Genome Biol Evol. 2013;5(3):599-605. doi: 10.1093/gbe/evt033. Genome Biol Evol. 2013. PMID: 23475937 Free PMC article.
-
Old Trade, New Tricks: Insights into the Spontaneous Mutation Process from the Partnering of Classical Mutation Accumulation Experiments with High-Throughput Genomic Approaches.Genome Biol Evol. 2019 Jan 1;11(1):136-165. doi: 10.1093/gbe/evy252. Genome Biol Evol. 2019. PMID: 30476040 Free PMC article. Review.
-
Analysis and implications of mutational variation.Genetica. 2009 Jun;136(2):359-69. doi: 10.1007/s10709-008-9304-4. Epub 2008 Jul 29. Genetica. 2009. PMID: 18663587 Review.
Cited by
-
The Rate and Molecular Spectrum of Spontaneous Mutations in the GC-Rich Multichromosome Genome of Burkholderia cenocepacia.Genetics. 2015 Jul;200(3):935-46. doi: 10.1534/genetics.115.176834. Epub 2015 May 12. Genetics. 2015. PMID: 25971664 Free PMC article.
-
Population Bottlenecks Strongly Influence the Evolutionary Trajectory to Fluoroquinolone Resistance in Escherichia coli.Mol Biol Evol. 2020 Jun 1;37(6):1637-1646. doi: 10.1093/molbev/msaa032. Mol Biol Evol. 2020. PMID: 32031639 Free PMC article.
-
Low mutational load and high mutation rate variation in gut commensal bacteria.PLoS Biol. 2020 Mar 10;18(3):e3000617. doi: 10.1371/journal.pbio.3000617. eCollection 2020 Mar. PLoS Biol. 2020. PMID: 32155146 Free PMC article.
-
The effect of induced mutations on quantitative traits in Arabidopsis thaliana: Natural versus artificial conditions.Ecol Evol. 2016 Oct 24;6(23):8366-8374. doi: 10.1002/ece3.2558. eCollection 2016 Dec. Ecol Evol. 2016. PMID: 28031789 Free PMC article.
-
Temperature responses of mutation rate and mutational spectrum in an Escherichia coli strain and the correlation with metabolic rate.BMC Evol Biol. 2018 Aug 29;18(1):126. doi: 10.1186/s12862-018-1252-8. BMC Evol Biol. 2018. PMID: 30157765 Free PMC article.
References
-
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247. - PubMed
-
- Bateman A., 1959. The viability of near-normal irradiated chromosomes. Int. J. Radiat. Biol. 1: 170–180.
-
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple Testing. J Roy Stat Soc B Met 57: 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

