Microbial glycan microarrays define key features of host-microbial interactions
- PMID: 24814672
- PMCID: PMC4158828
- DOI: 10.1038/nchembio.1525
Microbial glycan microarrays define key features of host-microbial interactions
Abstract
Genomic approaches continue to provide unprecedented insight into the microbiome, yet host immune interactions with diverse microbiota can be difficult to study. We therefore generated a microbial microarray containing defined antigens isolated from a broad range of microbial flora to examine adaptive and innate immunity. Serological studies with this microarray show that immunoglobulins from multiple mammalian species have unique patterns of reactivity, whereas exposure of animals to distinct microbes induces specific serological recognition. Although adaptive immunity exhibited plasticity toward microbial antigens, immunological tolerance limits reactivity toward self. We discovered that several innate immune galectins show specific recognition of microbes that express self-like antigens, leading to direct killing of a broad range of Gram-negative and Gram-positive microbes. Thus, host protection against microbes seems to represent a balance between adaptive and innate immunity to defend against evolving antigenic determinants while protecting against molecular mimicry.
Figures

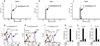
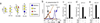
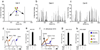


Comment in
-
Techniques and applications: Self-like antigens under fire.Nat Rev Microbiol. 2014 Jul;12(7):462-3. doi: 10.1038/nrmicro3290. Epub 2014 May 27. Nat Rev Microbiol. 2014. PMID: 24861038 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- P01HL107151/HL/NHLBI NIH HHS/United States
- P01 HL107151/HL/NHLBI NIH HHS/United States
- P01 HL085607/HL/NHLBI NIH HHS/United States
- U54GM62116/GM/NIGMS NIH HHS/United States
- P41 GM103694/GM/NIGMS NIH HHS/United States
- P41GM103694/GM/NIGMS NIH HHS/United States
- U54 GM062116/GM/NIGMS NIH HHS/United States
- T32 HL069769/HL/NHLBI NIH HHS/United States
- R24 GM098791/GM/NIGMS NIH HHS/United States
- R01 AI050143/AI/NIAID NIH HHS/United States
- R24GM098791/GM/NIGMS NIH HHS/United States
- R01AI050143/AI/NIAID NIH HHS/United States
- HL085607/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

