Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota
- PMID: 24814785
- PMCID: PMC4068677
- DOI: 10.1128/AEM.00079-14
Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota
Abstract
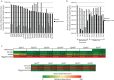
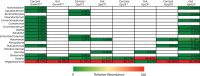
The invertebrate microbiome contributes to multiple aspects of host physiology, including nutrient supplementation and immune maturation processes. We identified and compared gut microbial abundance and diversity in natural tsetse flies from Uganda using five genetically distinct populations of Glossina fuscipes fuscipes and multiple tsetse species (Glossina morsitans morsitans, G. f. fuscipes, and Glossina pallidipes) that occur in sympatry in one location. We used multiple approaches, including deep sequencing of the V4 hypervariable region of the 16S rRNA gene, 16S rRNA gene clone libraries, and bacterium-specific quantitative PCR (qPCR), to investigate the levels and patterns of gut microbial diversity from a total of 151 individuals. Our results show extremely limited diversity in field flies of different tsetse species. The obligate endosymbiont Wigglesworthia dominated all samples (>99%), but we also observed wide prevalence of low-density Sodalis (tsetse's commensal endosymbiont) infections (<0.05%). There were also several individuals (22%) with high Sodalis density, which also carried coinfections with Serratia. Albeit in low density, we noted differences in microbiota composition among the genetically distinct G. f. fuscipes flies and between different sympatric species. Interestingly, Wigglesworthia density varied in different species (10(4) to 10(6) normalized genomes), with G. f. fuscipes having the highest levels. We describe the factors that may be responsible for the reduced diversity of tsetse's gut microbiota compared to those of other insects. Additionally, we discuss the implications of Wigglesworthia and Sodalis density variations as they relate to trypanosome transmission dynamics and vector competence variations associated with different tsetse species.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

