Flexible torsion-angle noncrystallographic symmetry restraints for improved macromolecular structure refinement
- PMID: 24816103
- PMCID: PMC4014122
- DOI: 10.1107/S1399004714003277
Flexible torsion-angle noncrystallographic symmetry restraints for improved macromolecular structure refinement
Abstract
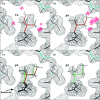
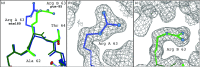
One of the great challenges in refining macromolecular crystal structures is a low data-to-parameter ratio. Historically, knowledge from chemistry has been used to help to improve this ratio. When a macromolecule crystallizes with more than one copy in the asymmetric unit, the noncrystallographic symmetry relationships can be exploited to provide additional restraints when refining the working model. However, although globally similar, NCS-related chains often have local differences. To allow for local differences between NCS-related molecules, flexible torsion-based NCS restraints have been introduced, coupled with intelligent rotamer handling for protein chains, and are available in phenix.refine for refinement of models at all resolutions.
Keywords: NCS; automation; macromolecular crystallography; noncrystallographic symmetry; refinement.
Figures





Similar articles
-
Accurate macromolecular crystallographic refinement: incorporation of the linear scaling, semiempirical quantum-mechanics program DivCon into the PHENIX refinement package.Acta Crystallogr D Biol Crystallogr. 2014 May;70(Pt 5):1233-47. doi: 10.1107/S1399004714002260. Epub 2014 Apr 26. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24816093 Free PMC article.
-
Real-space refinement in PHENIX for cryo-EM and crystallography.Acta Crystallogr D Struct Biol. 2018 Jun 1;74(Pt 6):531-544. doi: 10.1107/S2059798318006551. Epub 2018 May 30. Acta Crystallogr D Struct Biol. 2018. PMID: 29872004 Free PMC article.
-
Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution.Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):381-90. doi: 10.1107/S0907444911047834. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505258 Free PMC article.
-
Keep it together: restraints in crystallographic refinement of macromolecule-ligand complexes.Acta Crystallogr D Struct Biol. 2017 Feb 1;73(Pt 2):93-102. doi: 10.1107/S2059798316017964. Epub 2017 Feb 1. Acta Crystallogr D Struct Biol. 2017. PMID: 28177305 Free PMC article. Review.
-
Recent developments for the efficient crystallographic refinement of macromolecular structures.Curr Opin Struct Biol. 1998 Oct;8(5):606-11. doi: 10.1016/s0959-440x(98)80152-8. Curr Opin Struct Biol. 1998. PMID: 9818265 Review.
Cited by
-
The cAMP receptor protein from Gardnerella vaginalis is not regulated by ligands.Commun Biol. 2024 Oct 1;7(1):1233. doi: 10.1038/s42003-024-06957-1. Commun Biol. 2024. PMID: 39354127 Free PMC article.
-
De novo design of buttressed loops for sculpting protein functions.Nat Chem Biol. 2024 Aug;20(8):974-980. doi: 10.1038/s41589-024-01632-2. Epub 2024 May 30. Nat Chem Biol. 2024. PMID: 38816644 Free PMC article.
-
Design of protein-binding proteins from the target structure alone.Nature. 2022 May;605(7910):551-560. doi: 10.1038/s41586-022-04654-9. Epub 2022 Mar 24. Nature. 2022. PMID: 35332283 Free PMC article.
-
Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix.Acta Crystallogr D Struct Biol. 2019 Oct 1;75(Pt 10):861-877. doi: 10.1107/S2059798319011471. Epub 2019 Oct 2. Acta Crystallogr D Struct Biol. 2019. PMID: 31588918 Free PMC article.
-
In situ ligand restraints from quantum-mechanical methods.Acta Crystallogr D Struct Biol. 2023 Feb 1;79(Pt 2):100-110. doi: 10.1107/S2059798323000025. Epub 2023 Jan 20. Acta Crystallogr D Struct Biol. 2023. PMID: 36762856 Free PMC article.
References
-
- Bricogne, G. (1976). Acta Cryst. A32, 832–847.
-
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. & Vonrhein, C. (2010). BUSTER. Global Phasing Limited, Cambridge, England.
-
- Brünger, A. T. (1992). Nature (London), 355, 472–475. - PubMed
-
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

