An arginine tetrad as mediator of input-dependent and input-independent ATPases in the clock protein KaiC
- PMID: 24816106
- PMCID: PMC4722857
- DOI: 10.1107/S1399004714003228
An arginine tetrad as mediator of input-dependent and input-independent ATPases in the clock protein KaiC
Abstract
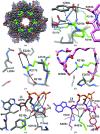
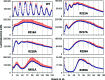
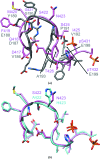
A post-translational oscillator (PTO) composed of the proteins KaiA, KaiB and KaiC is at the heart of the cyanobacterial circadian clock. KaiC interacts with KaiA and KaiB over the daily cycle, and CII domains undergo rhythmic phosphorylation/dephosphorylation with a 24 h period. Both the N-terminal (CI) and C-terminal (CII) rings of KaiC exhibit ATPase activity. The CI ATPase proceeds in an input-independent fashion, but the CII ATPase is subject to metabolic input signals. The crystal structure of KaiC from Thermosynechococcus elongatus allows insight into the different anatomies of the CI and CII ATPases. Four consecutive arginines in CI (Arg linker) that connect the P-loop, CI subunits and CI and CII at the ring interface are primary candidates for the coordination of the CI and CII activities. The mutation of linker residues alters the period or triggers arhythmic behavior. Comparison between the CI and CII structures also reveals differences in loop regions that are key to KaiA and KaiB binding and activation of CII ATPase and kinase. Common packing features in KaiC crystals shed light on the KaiB-KaiC interaction.
Keywords: ATPase; KaiABC; circadian clock; kinase.
Figures






Similar articles
-
Architecture and mechanism of the central gear in an ancient molecular timer.J R Soc Interface. 2017 Mar;14(128):20161065. doi: 10.1098/rsif.2016.1065. J R Soc Interface. 2017. PMID: 28330987 Free PMC article. Review.
-
Intricate protein-protein interactions in the cyanobacterial circadian clock.J Biol Chem. 2014 Aug 1;289(31):21267-75. doi: 10.1074/jbc.R114.579607. Epub 2014 Jun 16. J Biol Chem. 2014. PMID: 24936066 Free PMC article. Review.
-
Nature of KaiB-KaiC binding in the cyanobacterial circadian oscillator.Cell Cycle. 2013 Mar 1;12(5):810-7. doi: 10.4161/cc.23757. Epub 2013 Feb 6. Cell Cycle. 2013. PMID: 23388462 Free PMC article.
-
Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism.Biochemistry. 2012 Feb 28;51(8):1547-58. doi: 10.1021/bi201525n. Epub 2012 Feb 13. Biochemistry. 2012. PMID: 22304631 Free PMC article.
-
Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase.PLoS One. 2009 Nov 26;4(11):e7529. doi: 10.1371/journal.pone.0007529. PLoS One. 2009. PMID: 19956664 Free PMC article.
Cited by
-
Architecture and mechanism of the central gear in an ancient molecular timer.J R Soc Interface. 2017 Mar;14(128):20161065. doi: 10.1098/rsif.2016.1065. J R Soc Interface. 2017. PMID: 28330987 Free PMC article. Review.
-
Elucidation of master allostery essential for circadian clock oscillation in cyanobacteria.Sci Adv. 2022 Apr 15;8(15):eabm8990. doi: 10.1126/sciadv.abm8990. Epub 2022 Apr 15. Sci Adv. 2022. PMID: 35427168 Free PMC article.
-
Coupling of distant ATPase domains in the circadian clock protein KaiC.Nat Struct Mol Biol. 2022 Aug;29(8):759-766. doi: 10.1038/s41594-022-00803-w. Epub 2022 Jul 21. Nat Struct Mol Biol. 2022. PMID: 35864165 Free PMC article.
-
Intricate protein-protein interactions in the cyanobacterial circadian clock.J Biol Chem. 2014 Aug 1;289(31):21267-75. doi: 10.1074/jbc.R114.579607. Epub 2014 Jun 16. J Biol Chem. 2014. PMID: 24936066 Free PMC article. Review.
-
Minimal tool set for a prokaryotic circadian clock.BMC Evol Biol. 2017 Jul 21;17(1):169. doi: 10.1186/s12862-017-0999-7. BMC Evol Biol. 2017. PMID: 28732467 Free PMC article.
References
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221. - PubMed
-
- Aronson, B. D., Johnson, K. A., Loros, J. J. & Dunlap, J. C. (1994). Science, 263, 1578–1584. - PubMed
-
- Brenner, S. L., Mitchell, R. S., Morrical, S. W., Neuendorf, S. K., Schutte, B. C. & Cox, M. M. (1987). J. Biol. Chem. 262, 4011–4016. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

