Fc receptors for immunoglobulins and their appearance during vertebrate evolution
- PMID: 24816777
- PMCID: PMC4016189
- DOI: 10.1371/journal.pone.0096903
Fc receptors for immunoglobulins and their appearance during vertebrate evolution
Erratum in
-
Correction: Fc receptors for immunoglobulins and their appearance during vertebrate evolution.PLoS One. 2015 Apr 1;10(4):e0124530. doi: 10.1371/journal.pone.0124530. eCollection 2015. PLoS One. 2015. PMID: 25830328 Free PMC article. No abstract available.
Abstract
Receptors interacting with the constant domain of immunoglobulins (Igs) have a number of important functions in vertebrates. They facilitate phagocytosis by opsonization, are key components in antibody-dependent cellular cytotoxicity as well as activating cells to release granules. In mammals, four major types of classical Fc receptors (FcRs) for IgG have been identified, one high-affinity receptor for IgE, one for both IgM and IgA, one for IgM and one for IgA. All of these receptors are related in structure and all of them, except the IgA receptor, are found in primates on chromosome 1, indicating that they originate from a common ancestor by successive gene duplications. The number of Ig isotypes has increased gradually during vertebrate evolution and this increase has likely been accompanied by a similar increase in isotype-specific receptors. To test this hypothesis we have performed a detailed bioinformatics analysis of a panel of vertebrate genomes. The first components to appear are the poly-Ig receptors (PIGRs), receptors similar to the classic FcRs in mammals, so called FcRL receptors, and the FcR γ chain. These molecules are not found in cartilagous fish and may first appear within bony fishes, indicating a major step in Fc receptor evolution at the appearance of bony fish. In contrast, the receptor for IgA is only found in placental mammals, indicating a relatively late appearance. The IgM and IgA/M receptors are first observed in the monotremes, exemplified by the platypus, indicating an appearance during early mammalian evolution. Clearly identifiable classical receptors for IgG and IgE are found only in marsupials and placental mammals, but closely related receptors are found in the platypus, indicating a second major step in Fc receptor evolution during early mammalian evolution, involving the appearance of classical IgG and IgE receptors from FcRL molecules and IgM and IgA/M receptors from PIGR.
Conflict of interest statement
Figures


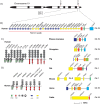

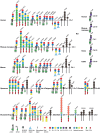
References
-
- Zhao Y, Cui H, Whittington CM, Wei Z, Zhang X, et al. (2009) Ornithorhynchus anatinus (platypus) links the evolution of immunoglobulin genes in eutherian mammals and nonmammalian tetrapods. J Immunol 183: 3285–3293. - PubMed
-
- Dooley H, Flajnik MF (2006) Antibody repertoire development in cartilaginous fish. Dev Comp Immunol 30: 43–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

