OTX2 duplication is implicated in hemifacial microsomia
- PMID: 24816892
- PMCID: PMC4016008
- DOI: 10.1371/journal.pone.0096788
OTX2 duplication is implicated in hemifacial microsomia
Abstract
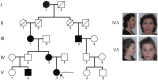
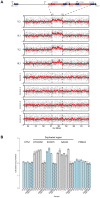
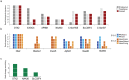
Hemifacial microsomia (HFM) is the second most common facial anomaly after cleft lip and palate. The phenotype is highly variable and most cases are sporadic. We investigated the disorder in a large pedigree with five affected individuals spanning eight meioses. Whole-exome sequencing results indicated the absence of a pathogenic coding point mutation. A genome-wide survey of segmental variations identified a 1.3 Mb duplication of chromosome 14q22.3 in all affected individuals that was absent in more than 1000 chromosomes of ethnically matched controls. The duplication was absent in seven additional sporadic HFM cases, which is consistent with the known heterogeneity of the disorder. To find the critical gene in the duplicated region, we analyzed signatures of human craniofacial disease networks, mouse expression data, and predictions of dosage sensitivity. All of these approaches implicated OTX2 as the most likely causal gene. Moreover, OTX2 is a known oncogenic driver in medulloblastoma, a condition that was diagnosed in the proband during the course of the study. Our findings suggest a role for OTX2 dosage sensitivity in human craniofacial development and raise the possibility of a shared etiology between a subtype of hemifacial microsomia and medulloblastoma.
Conflict of interest statement
Figures
References
-
- Gorlin RJ CM, Hennekam RCM (2001) Syndromes of the head and neck. New York: Oxford University Press.
-
- Heike CL, Hing AV (2009) Craniofacial Microsomia Overview. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, et al.., editors. GeneReviews. Seattle (WA).
-
- Rimoin DL EA (2006) Emery and Rimoin's principles and practice of medical genetics. Philadelphia: Churchill Livingstone.
-
- Poswillo D (1988) The aetiology and pathogenesis of craniofacial deformity. Development 103 Suppl 207–212. - PubMed
-
- Werler MM, Sheehan JE, Hayes C, Padwa BL, Mitchell AA, et al. (2004) Demographic and reproductive factors associated with hemifacial microsomia. Cleft Palate Craniofac J 41: 494–450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

