Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum
- PMID: 24817274
- PMCID: PMC4016276
- DOI: 10.1371/journal.pone.0097148
Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum
Abstract
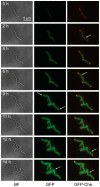
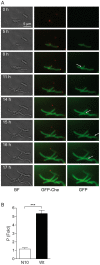
Understanding the mechanism that controls space-time coordination of elongation and division of Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is critical for fighting the tubercle bacillus. Most of the numerous enzymes involved in the synthesis of Mycolic acid - Arabinogalactan-Peptidoglycan complex (MAPc) in the cell wall are essential in vivo. Using a dynamic approach, we localized Mtb enzymes belonging to the fatty acid synthase-II (FAS-II) complexes and involved in mycolic acid (MA) biosynthesis in a mycobacterial model of Mtb: M. smegmatis. Results also showed that the MA transporter MmpL3 was present in the mycobacterial envelope and was specifically and dynamically accumulated at the poles and septa during bacterial growth. This localization was due to its C-terminal domain. Moreover, the FAS-II enzymes were co-localized at the poles and septum with Wag31, the protein responsible for the polar localization of mycobacterial peptidoglycan biosynthesis. The dynamic localization of FAS-II and of the MA transporter with Wag31, at the old-growing poles and at the septum suggests that the main components of the mycomembrane may potentially be synthesized at these precise foci. This finding highlights a major difference between mycobacteria and other rod-shaped bacteria studied to date. Based on the already known polar activities of envelope biosynthesis in mycobacteria, we propose the existence of complex polar machinery devoted to the biogenesis of the entire envelope. As a result, the mycobacterial pole would represent the Achilles' heel of the bacillus at all its growing stages.
Conflict of interest statement
Figures


References
-
- Daffe M, Draper P (1998) The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol 39: 131–203. - PubMed
-
- Minnikin DE, Kremer L, Dover LG, Besra GS (2002) The methyl-branched fortifications of Mycobacterium tuberculosis. Chem Biol 9: 545–553. - PubMed
-
- Ernst JD (2012) The immunological life cycle of tuberculosis. Nat Rev Immunol 12: 581–591. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

