bZIPs and WRKYs: two large transcription factor families executing two different functional strategies
- PMID: 24817872
- PMCID: PMC4012195
- DOI: 10.3389/fpls.2014.00169
bZIPs and WRKYs: two large transcription factor families executing two different functional strategies
Abstract
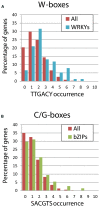
bZIPs and WRKYs are two important plant transcription factor (TF) families regulating diverse developmental and stress-related processes. Since a partial overlap in these biological processes is obvious, it can be speculated that they fulfill non-redundant functions in a complex regulatory network. Here, we focus on the regulatory mechanisms that are so far described for bZIPs and WRKYs. bZIP factors need to heterodimerize for DNA-binding and regulation of transcription, and based on a bioinformatics approach, bZIPs can build up more than the double of protein interactions than WRKYs. In contrast, an enrichment of the WRKY DNA-binding motifs can be found in WRKY promoters, a phenomenon which is not observed for the bZIP family. Thus, the two TF families follow two different functional strategies in which WRKYs regulate each other's transcription in a transcriptional network whereas bZIP action relies on intensive heterodimerization.
Keywords: DNA-binding; G/C box accumulation; W-box accumulation; WRKYs; bZIPs; heterodimerization; regulatory mechanisms.
Figures



Similar articles
-
Transcription factor ThWRKY4 binds to a novel WLS motif and a RAV1A element in addition to the W-box to regulate gene expression.Plant Sci. 2017 Aug;261:38-49. doi: 10.1016/j.plantsci.2017.04.016. Epub 2017 May 4. Plant Sci. 2017. PMID: 28554692
-
WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants.Front Plant Sci. 2016 Jun 3;7:760. doi: 10.3389/fpls.2016.00760. eCollection 2016. Front Plant Sci. 2016. PMID: 27375634 Free PMC article. Review.
-
Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress.Plant Cell. 2011 Jan;23(1):381-95. doi: 10.1105/tpc.110.075390. Epub 2011 Jan 28. Plant Cell. 2011. PMID: 21278122 Free PMC article.
-
Phylogenetic and transcriptional analysis of an expanded bZIP transcription factor family in Phytophthora sojae.BMC Genomics. 2013 Nov 28;14(1):839. doi: 10.1186/1471-2164-14-839. BMC Genomics. 2013. PMID: 24286285 Free PMC article.
-
Basic leucine zipper domain transcription factors: the vanguards in plant immunity.Biotechnol Lett. 2017 Dec;39(12):1779-1791. doi: 10.1007/s10529-017-2431-1. Epub 2017 Sep 6. Biotechnol Lett. 2017. PMID: 28879532 Review.
Cited by
-
Dynamic subnuclear relocalisation of WRKY40 in response to Abscisic acid in Arabidopsis thaliana.Sci Rep. 2015 Aug 21;5:13369. doi: 10.1038/srep13369. Sci Rep. 2015. PMID: 26293691 Free PMC article.
-
Coiled-coil motif in LBD16 and LBD18 transcription factors are critical for dimerization and biological function in arabidopsis.Plant Signal Behav. 2018 Jan 2;13(1):e1411450. doi: 10.1080/15592324.2017.1411450. Epub 2017 Dec 21. Plant Signal Behav. 2018. PMID: 29227192 Free PMC article.
-
Genome wide identification of LcC2DPs gene family in Lotus corniculatus provides insights into regulatory network in response to abiotic stresses.Sci Rep. 2025 Apr 18;15(1):13380. doi: 10.1038/s41598-025-97896-2. Sci Rep. 2025. PMID: 40251318 Free PMC article.
-
The phylogeny of C/S1 bZIP transcription factors reveals a shared algal ancestry and the pre-angiosperm translational regulation of S1 transcripts.Sci Rep. 2016 Jul 26;6:30444. doi: 10.1038/srep30444. Sci Rep. 2016. PMID: 27457880 Free PMC article.
-
Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions.Front Plant Sci. 2016 Jul 14;7:1029. doi: 10.3389/fpls.2016.01029. eCollection 2016. Front Plant Sci. 2016. PMID: 27471513 Free PMC article. Review.
References
-
- Alonso R., Onate-Sanchez L., Weltmeier F., Ehlert A., Diaz I., Dietrich K., et al. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21 1747–1761 10.1105/tpc.108.062968 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

