L1 retrotransposition: The snap-velcro model and its consequences
- PMID: 24818067
- PMCID: PMC4014453
- DOI: 10.4161/mge.28907
L1 retrotransposition: The snap-velcro model and its consequences
Abstract
LINE-1 (L1) elements are the only active and autonomous transposable elements in humans. The core retrotransposition machinery is a ribonucleoprotein particle (RNP) containing the L1 mRNA, with endonuclease and reverse transcriptase activities. It initiates reverse transcription directly at genomic target sites upon endonuclease cleavage. Recently, using a direct L1 extension assay (DLEA), we systematically tested the ability of native L1 RNPs to extend DNA substrates of various sequences and structures. We deduced from these experiments the general rules guiding the initiation of L1 reverse transcription, referred to as the snap-velcro model. In this model, L1 target choice is not only mediated by the sequence specificity of the endonuclease, but also through base-pairing between the L1 mRNA and the target site, which permits the subsequent L1 reverse transcription step. In addition, L1 reverse transcriptase efficiently primes L1 DNA synthesis only when the 3' end of the DNA substrate is single-stranded, suggesting so-far unrecognized DNA processing steps at the integration site.
Keywords: L1; LINE-1; TPRT; endonuclease; non-LTR retrotransposon; poly(A) tail; reverse transcriptase; reverse transcription; target-primed reverse transcription.
Figures
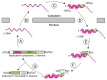
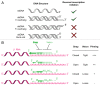

Comment on
- Monot C, Kuciak M, Viollet S, Mir AA, Gabus C, Darlix JL, Cristofari G. The specificity and flexibility of l1 reverse transcription priming at imperfect T-tracts. PLoS Genet. 2013;9:e1003499. doi: 10.1371/journal.pgen.1003499.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
