Detection of food spoilage and pathogenic bacteria based on ligation detection reaction coupled to flow-through hybridization on membranes
- PMID: 24818128
- PMCID: PMC4004135
- DOI: 10.1155/2014/156323
Detection of food spoilage and pathogenic bacteria based on ligation detection reaction coupled to flow-through hybridization on membranes
Abstract
Traditional culturing methods are still commonly applied for bacterial identification in the food control sector, despite being time and labor intensive. Microarray technologies represent an interesting alternative. However, they require higher costs and technical expertise, making them still inappropriate for microbial routine analysis. The present study describes the development of an efficient method for bacterial identification based on flow-through reverse dot-blot (FT-RDB) hybridization on membranes, coupled to the high specific ligation detection reaction (LDR). First, the methodology was optimized by testing different types of ligase enzymes, labeling, and membranes. Furthermore, specific oligonucleotide probes were designed based on the 16S rRNA gene, using the bioinformatic tool Oligonucleotide Retrieving for Molecular Applications (ORMA). Four probes were selected and synthesized, being specific for Aeromonas spp., Pseudomonas spp., Shewanella spp., and Morganella morganii, respectively. For the validation of the probes, 16 reference strains from type culture collections were tested by LDR and FT-RDB hybridization using universal arrays spotted onto membranes. In conclusion, the described methodology could be applied for the rapid, accurate, and cost-effective identification of bacterial species, exhibiting special relevance in food safety and quality.
Figures



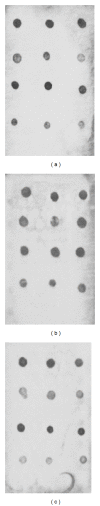

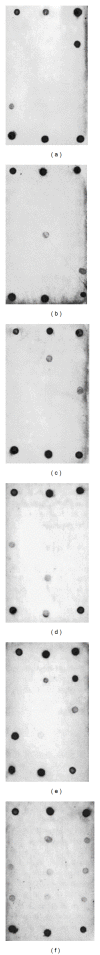
Similar articles
-
High-throughput detection of food-borne pathogenic bacteria using oligonucleotide microarray with quantum dots as fluorescent labels.Int J Food Microbiol. 2014 Aug 18;185:27-32. doi: 10.1016/j.ijfoodmicro.2014.05.012. Epub 2014 May 21. Int J Food Microbiol. 2014. PMID: 24927399
-
Rapid, species-specific detection of uropathogen 16S rDNA and rRNA at ambient temperature by dot-blot hybridization and an electrochemical sensor array.Mol Genet Metab. 2005 Jan;84(1):90-9. doi: 10.1016/j.ymgme.2004.11.006. Epub 2004 Dec 13. Mol Genet Metab. 2005. PMID: 15639199
-
Comparison of real-time PCR with SYBR Green I or 5'-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria.Microbiology (Reading). 2003 Jan;149(Pt 1):269-77. doi: 10.1099/mic.0.25975-0. Microbiology (Reading). 2003. PMID: 12576600
-
[Studies on rapid detection of food-borne pathogenic bacteria by nucleic acid testing and related technology].Wei Sheng Yan Jiu. 2008 Mar;37(2):245-8. Wei Sheng Yan Jiu. 2008. PMID: 18589620 Review. Chinese.
-
Detection of pathogenic bacteria in foods by using recombinant DNA techniques.Bioprocess Technol. 1994;19:605-19. Bioprocess Technol. 1994. PMID: 7764778 Review. No abstract available.
Cited by
-
A novel fluorescent biosensor based on dendritic DNA nanostructure in combination with ligase reaction for ultrasensitive detection of DNA methylation.J Nanobiotechnology. 2019 Dec 7;17(1):121. doi: 10.1186/s12951-019-0552-5. J Nanobiotechnology. 2019. PMID: 31812164 Free PMC article.
-
Fluorescence-based bioassays for the detection and evaluation of food materials.Sensors (Basel). 2015 Oct 13;15(10):25831-67. doi: 10.3390/s151025831. Sensors (Basel). 2015. PMID: 26473869 Free PMC article. Review.
-
IoT-Enabled Electronic Nose System for Beef Quality Monitoring and Spoilage Detection.Foods. 2023 May 31;12(11):2227. doi: 10.3390/foods12112227. Foods. 2023. PMID: 37297471 Free PMC article.
References
-
- Zhou J. Microarrays for bacterial detection and microbial community analysis. Current Opinion in Microbiology. 2003;6(3):288–294. - PubMed
-
- Gentry TJ, Zhou J. Microarray-based microbial identification and characterization. In: Tang Y-W, Stratton CW, editors. Advanced Techniques in Diagnostic Microbiology. New York, NY, USA: Springer; 2006. pp. 276–290.
-
- Wang X-W, Zhang L, Jin L-Q, et al. Development and application of an oligonucleotide microarray for the detection of food-borne bacterial pathogens. Applied Microbiology and Biotechnology. 2007;76(1):225–233. - PubMed
-
- Eom HS, Hwang BH, Kim D-H, Lee I-B, Kim YH, Cha HJ. Multiple detection of food-borne pathogenic bacteria using a novel 16S rDNA-based oligonucleotide signature chip. Biosensors and Bioelectronics. 2007;22(6):845–853. - PubMed
-
- Stine OC, Carnahan A, Singh R, et al. Characterization of microbial communities from coastal waters using microarrays. Environmental Monitoring and Assessment. 2003;81(1–3):327–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

