Chd5 orchestrates chromatin remodelling during sperm development
- PMID: 24818823
- PMCID: PMC4151132
- DOI: 10.1038/ncomms4812
Chd5 orchestrates chromatin remodelling during sperm development
Abstract
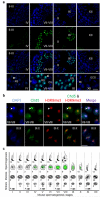
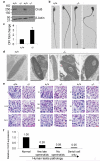
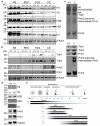
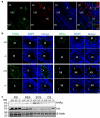
One of the most remarkable chromatin remodelling processes occurs during spermiogenesis, the post-meiotic phase of sperm development during which histones are replaced with sperm-specific protamines to repackage the genome into the highly compact chromatin structure of mature sperm. Here we identify Chromodomain helicase DNA binding protein 5 (Chd5) as a master regulator of the histone-to-protamine chromatin remodelling process. Chd5 deficiency leads to defective sperm chromatin compaction and male infertility in mice, mirroring the observation of low CHD5 expression in testes of infertile men. Chd5 orchestrates a cascade of molecular events required for histone removal and replacement, including histone 4 (H4) hyperacetylation, histone variant expression, nucleosome eviction and DNA damage repair. Chd5 deficiency also perturbs expression of transition proteins (Tnp1/Tnp2) and protamines (Prm1/2). These findings define Chd5 as a multi-faceted mediator of histone-to-protamine replacement and depict the cascade of molecular events underlying this process of extensive chromatin remodelling.
Figures
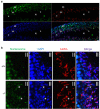
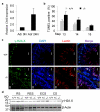
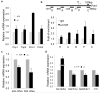
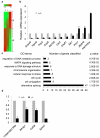
Comment in
-
Packing for the journey: CHD5 remodels the genome.Cell Cycle. 2014;13(12):1833-4. doi: 10.4161/cc.29378. Epub 2014 May 27. Cell Cycle. 2014. PMID: 24865185 Free PMC article. No abstract available.
References
-
- Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Advances in experimental medicine and biology. 2008;636:1–15. doi:10.1007/978-0-387-09597-4_1. - PubMed
-
- Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. The American journal of anatomy. 1956;99:507–516. doi:10.1002/aja.1000990307. - PubMed
-
- Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. The American journal of anatomy. 1956;99:391–413. doi:10.1002/aja.1000990303. - PubMed
-
- Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods in molecular biology. 2009;558:263–277. doi:10.1007/978-1-60761-103-5_16. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

