Zinc finger endonuclease targeting PSIP1 inhibits HIV-1 integration
- PMID: 24820090
- PMCID: PMC4136077
- DOI: 10.1128/AAC.02690-14
Zinc finger endonuclease targeting PSIP1 inhibits HIV-1 integration
Abstract
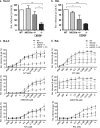
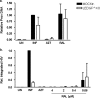
Genome editing using zinc finger nucleases (ZFNs) has been successfully applied to disrupt CCR5 or CXCR4 host factors and inhibit viral entry and infection. Gene therapy using ZFNs to modify the PSIP1 gene, which encodes the lens epithelium-derived growth factor (LEDGF) protein, might restrain an early step of the viral replication cycle at the integration level. ZFNs targeting the PSIP1 gene (ZFNLEDGF) were designed to specifically recognize the sequence after the integrase binding domain (IBD) of the LEDGF/p75 protein. ZFNLEDGF successfully recognized the target region of the PSIP1 gene in TZM-bl cells by heteroduplex formation and DNA sequence analysis. Gene editing induced a frameshift of the coding region and resulted in the abolishment of LEDGF expression at the mRNA and protein levels. Functional assays revealed that infection with the HIV-1 R5 BaL or X4 NL4-3 viral strains was impaired in LEDGF/p75 knockout cells regardless of entry tropism due to a blockade in HIV-1 proviral integration into the host genome. However, residual infection was detected in the LEDGF knockout cells. Indeed, LEDGF knockout restriction was overcome at a high multiplicity of infection, suggesting alternative mechanisms for HIV-1 genome integration rather than through LEDGF/p75. However, the observed residual integration was sensitive to the integrase inhibitor raltegravir. These results demonstrate that the described ZFNLEDGF effectively targets the PSIP1 gene, which is involved in the early steps of the viral replication cycle; thus, ZFNLEDGF may become a potential antiviral agent for restricting HIV-1 integration. Moreover, LEDGF knockout cells represent a potent tool for elucidating the role of HIV integration cofactors in virus replication.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism.J Virol. 2014 Sep 1;88(17):9704-17. doi: 10.1128/JVI.01397-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942577 Free PMC article.
-
Targeted editing of the PSIP1 gene encoding LEDGF/p75 protects cells against HIV infection.Sci Rep. 2019 Feb 20;9(1):2389. doi: 10.1038/s41598-019-38718-0. Sci Rep. 2019. PMID: 30787394 Free PMC article.
-
Peptides derived from the HIV-1 integrase promote HIV-1 infection and multi-integration of viral cDNA in LEDGF/p75-knockdown cells.Virol J. 2010 Aug 2;7:177. doi: 10.1186/1743-422X-7-177. Virol J. 2010. PMID: 20678206 Free PMC article.
-
Host factors for retroviral integration site selection.Trends Biochem Sci. 2015 Feb;40(2):108-16. doi: 10.1016/j.tibs.2014.12.001. Epub 2014 Dec 31. Trends Biochem Sci. 2015. PMID: 25555456 Review.
-
The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy.Virology. 2013 Jan 5;435(1):102-9. doi: 10.1016/j.virol.2012.09.033. Virology. 2013. PMID: 23217620 Review.
Cited by
-
Synthetic modified messenger RNA for therapeutic applications.Acta Biomater. 2021 Sep 1;131:1-15. doi: 10.1016/j.actbio.2021.06.020. Epub 2021 Jun 13. Acta Biomater. 2021. PMID: 34133982 Free PMC article. Review.
-
Nanomaterials for mRNA-based therapeutics: Challenges and opportunities.Bioeng Transl Med. 2023 Jan 29;8(3):e10492. doi: 10.1002/btm2.10492. eCollection 2023 May. Bioeng Transl Med. 2023. PMID: 37206219 Free PMC article. Review.
-
Genome-editing Technologies for Gene and Cell Therapy.Mol Ther. 2016 Mar;24(3):430-46. doi: 10.1038/mt.2016.10. Epub 2016 Jan 12. Mol Ther. 2016. PMID: 26755333 Free PMC article. Review.
-
Targeted gene exchange in plant cells mediated by a zinc finger nuclease double cut.Plant Biotechnol J. 2016 Apr;14(4):1151-60. doi: 10.1111/pbi.12483. Epub 2015 Oct 1. Plant Biotechnol J. 2016. PMID: 26426390 Free PMC article.
-
Bone Marrow Gene Therapy for HIV/AIDS.Viruses. 2015 Jul 17;7(7):3910-36. doi: 10.3390/v7072804. Viruses. 2015. PMID: 26193303 Free PMC article. Review.
References
-
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. 2010. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29:143–148. 10.1038/nbt.1755 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

