The Drosophila Su(var)3-7 gene is required for oogenesis and female fertility, genetically interacts with piwi and aubergine, but impacts only weakly transposon silencing
- PMID: 24820312
- PMCID: PMC4018442
- DOI: 10.1371/journal.pone.0096802
The Drosophila Su(var)3-7 gene is required for oogenesis and female fertility, genetically interacts with piwi and aubergine, but impacts only weakly transposon silencing
Abstract
Heterochromatin is made of repetitive sequences, mainly transposable elements (TEs), the regulation of which is critical for genome stability. We have analyzed the role of the heterochromatin-associated Su(var)3-7 protein in Drosophila ovaries. We present evidences that Su(var)3-7 is required for correct oogenesis and female fertility. It accumulates in heterochromatic domains of ovarian germline and somatic cells nuclei, where it co-localizes with HP1. Homozygous mutant females display ovaries with frequent degenerating egg-chambers. Absence of Su(var)3-7 in embryos leads to defects in meiosis and first mitotic divisions due to chromatin fragmentation or chromosome loss, showing that Su(var)3-7 is required for genome integrity. Females homozygous for Su(var)3-7 mutations strongly impair repression of P-transposable element induced gonadal dysgenesis but have minor effects on other TEs. Su(var)3-7 mutations reduce piRNA cluster transcription and slightly impact ovarian piRNA production. However, this modest piRNA reduction does not correlate with transposon de-silencing, suggesting that the moderate effect of Su(var)3-7 on some TE repression is not linked to piRNA production. Strikingly, Su(var)3-7 genetically interacts with the piwi and aubergine genes, key components of the piRNA pathway, by strongly impacting female fertility without impairing transposon silencing. These results lead us to propose that the interaction between Su(var)3-7 and piwi or aubergine controls important developmental processes independently of transposon silencing.
Conflict of interest statement
Figures

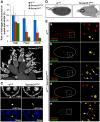
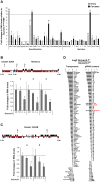
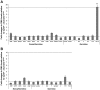
Similar articles
-
Direct interaction of Su(var)2-10 via the SIM-binding site of the Piwi protein is required for transposon silencing in Drosophila melanogaster.FEBS J. 2024 Apr;291(8):1759-1779. doi: 10.1111/febs.17073. Epub 2024 Feb 3. FEBS J. 2024. PMID: 38308815
-
Drosophila Modulo is essential for transposon silencing and developmental robustness.J Biol Chem. 2025 Mar;301(3):108210. doi: 10.1016/j.jbc.2025.108210. Epub 2025 Jan 22. J Biol Chem. 2025. PMID: 39848495 Free PMC article.
-
Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery.Genes Dev. 2015 Nov 1;29(21):2258-71. doi: 10.1101/gad.271908.115. Epub 2015 Oct 22. Genes Dev. 2015. PMID: 26494711 Free PMC article.
-
Untangling the web: the diverse functions of the PIWI/piRNA pathway.Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
-
Two distinct transcriptional controls triggered by nuclear Piwi-piRISCs in the Drosophila piRNA pathway.Curr Opin Struct Biol. 2018 Dec;53:69-76. doi: 10.1016/j.sbi.2018.06.005. Epub 2018 Jul 7. Curr Opin Struct Biol. 2018. PMID: 29990672 Review.
Cited by
-
P elements and the determinants of hybrid dysgenesis have different dynamics of propagation in Drosophila melanogaster populations.Genetica. 2015 Dec;143(6):751-9. doi: 10.1007/s10709-015-9872-z. Epub 2015 Nov 4. Genetica. 2015. PMID: 26530414
-
During a short window of Drosophila oogenesis, piRNA biogenesis may be boosted and mobilization of transposable elements allowed.Front Genet. 2014 Nov 6;5:385. doi: 10.3389/fgene.2014.00385. eCollection 2014. Front Genet. 2014. PMID: 25414724 Free PMC article. No abstract available.
References
-
- Delattre M, Spierer A, Tonka CH, Spierer P (2000) The genomic silencing of position-effect variegation in Drosophila melanogaster: interaction between the heterochromatin-associated proteins Su(var)3–7 and HP1. J Cell Sci 113 Pt 23: 4253–4261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

