Pericentromere tension is self-regulated by spindle structure in metaphase
- PMID: 24821839
- PMCID: PMC4018788
- DOI: 10.1083/jcb.201312024
Pericentromere tension is self-regulated by spindle structure in metaphase
Abstract
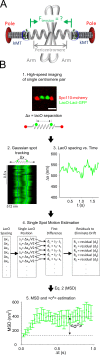
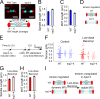
During cell division, a mitotic spindle is built by the cell and acts to align and stretch duplicated sister chromosomes before their ultimate segregation into daughter cells. Stretching of the pericentromeric chromatin during metaphase is thought to generate a tension-based signal that promotes proper chromosome segregation. However, it is not known whether the mitotic spindle actively maintains a set point tension magnitude for properly attached sister chromosomes to facilitate robust mechanochemical checkpoint signaling. By imaging and tracking the thermal movements of pericentromeric fluorescent markers in Saccharomyces cerevisiae, we measured pericentromere stiffness and then used the stiffness measurements to quantitatively evaluate the tension generated by pericentromere stretch during metaphase in wild-type cells and in mutants with disrupted chromosome structure. We found that pericentromere tension in yeast is substantial (4-6 pN) and is tightly self-regulated by the mitotic spindle: through adjustments in spindle structure, the cell maintains wild-type tension magnitudes even when pericentromere stiffness is disrupted.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

