Molecular genetics of clear-cell renal cell carcinoma
- PMID: 24821879
- PMCID: PMC4050206
- DOI: 10.1200/JCO.2012.45.2003
Molecular genetics of clear-cell renal cell carcinoma
Abstract
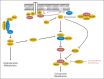
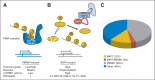
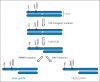
Renal cell carcinoma of clear-cell type (ccRCC) is an enigmatic tumor type, characterized by frequent inactivation of the VHL gene (infrequently mutated in other tumor types), responsiveness to angiogenesis inhibitors, and resistance to both chemotherapy and conventional radiation therapy. ccRCC tumors exhibit substantial mutation heterogeneity. Recent studies using massively parallel sequencing technologies have implicated several novel driver genes. In VHL wild-type tumors, mutations were discovered in TCEB1, which encodes Elongin C, a protein that binds to VHL and is required for its function. Several additional tumor suppressor genes have been identified near the VHL gene, within a region that is frequently deleted in ccRCC on chromosome 3p: SETD2, BAP1, and PBRM1. Mutations in BAP1 and PBRM1 are largely mutually exclusive and are associated with different tumor biology and patient outcomes. In addition, the mTORC1 pathway is deregulated by mutations in MTOR, TSC1, PIK3CA, and PTEN in approximately 20% of ccRCCs. Mutations in TSC1, and possibly other genes, may predict for sensitivity to mTORC1 inhibitors. These discoveries provide insight into ccRCC development and set the foundation for the first molecular genetic classification of the disease, paving the way for subtype-specific therapies.
© 2014 by American Society of Clinical Oncology.
Conflict of interest statement
Author's disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures
References
-
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. - PubMed
-
- Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2010. Bethesda, MD: National Cancer Institute; 2013.
-
- Baldewijns MM, van Vlodrop IJ, Schouten LJ, et al. Genetics and epigenetics of renal cell cancer. Biochim Biochim Biophys Acta. 2008;1785:133–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

