Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors
- PMID: 24822055
- PMCID: PMC4013460
- DOI: 10.3389/fgene.2014.00109
Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors
Abstract
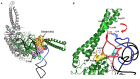
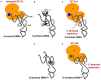
Conventional tRNAs have highly conserved sequences, four-armed cloverleaf secondary structures, and L-shaped tertiary structures. However, metazoan mitochondrial tRNAs contain several exceptional structures. Almost all tRNAs(Ser) for AGY/N codons lack the D-arm. Furthermore, in some nematodes, no four-armed cloverleaf-type tRNAs are present: two tRNAs(Ser) without the D-arm and 20 tRNAs without the T-arm are found. Previously, we showed that in nematode mitochondria, an extra elongation factor Tu (EF-Tu) has evolved to support interaction with tRNAs lacking the T-arm, which interact with C-terminal domain 3 in conventional EF-Tu. Recent mitochondrial genome analyses have suggested that in metazoan lineages other than nematodes, tRNAs without the T-arm are present. Furthermore, even more simplified tRNAs are predicted in some lineages. In this review, we discuss mitochondrial tRNAs with divergent structures, as well as protein factors, including EF-Tu, that support the function of truncated metazoan mitochondrial tRNAs.
Keywords: D-arm; T-arm; aminoacyl-tRNA synthetase; elongation factor Tu; mitochondrial tRNA; ribosome; tRNA nucleotidyltransferase.
Figures
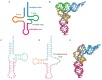
Similar articles
-
Duplication of Drosophila melanogaster mitochondrial EF-Tu: pre-adaptation to T-arm truncation and exclusion of bulky aminoacyl residues.Biochem J. 2017 Mar 7;474(6):957-969. doi: 10.1042/BCJ20160929. Biochem J. 2017. PMID: 28130490
-
An evolutionary 'intermediate state' of mitochondrial translation systems found in Trichinella species of parasitic nematodes: co-evolution of tRNA and EF-Tu.Nucleic Acids Res. 2006;34(18):5291-9. doi: 10.1093/nar/gkl526. Epub 2006 Sep 29. Nucleic Acids Res. 2006. PMID: 17012285 Free PMC article.
-
A protein extension to shorten RNA: elongated elongation factor-Tu recognizes the D-arm of T-armless tRNAs in nematode mitochondria.Biochem J. 2006 Oct 15;399(2):249-56. doi: 10.1042/BJ20060781. Biochem J. 2006. PMID: 16859488 Free PMC article.
-
T-armless tRNAs and elongated elongation factor Tu.IUBMB Life. 2007 Feb;59(2):68-75. doi: 10.1080/15216540701218722. IUBMB Life. 2007. PMID: 17454297 Review.
-
tRNA gene diversity in the three domains of life.Front Genet. 2014 May 26;5:142. doi: 10.3389/fgene.2014.00142. eCollection 2014. Front Genet. 2014. PMID: 24904642 Free PMC article. Review.
Cited by
-
The complete mitochondrial genome of the longneck croaker, pseudotolithus typus Bleeker, 1863 from Sierra Leone.Mitochondrial DNA B Resour. 2021 May 17;6(5):1640-1641. doi: 10.1080/23802359.2021.1927218. Mitochondrial DNA B Resour. 2021. PMID: 34104724 Free PMC article.
-
Novel insight into lepidopteran phylogenetics from the mitochondrial genome of the apple fruit moth of the family Argyresthiidae.BMC Genomics. 2024 Jan 2;25(1):21. doi: 10.1186/s12864-023-09905-1. BMC Genomics. 2024. PMID: 38166583 Free PMC article.
-
The complete mitochondrial genome of the Mexican-endemic cavefish Ophisternoninfernale (Synbranchiformes, Synbranchidae): insights on patterns of selection and implications for synbranchiform phylogenetics.Zookeys. 2022 Mar 11;1089:1-23. doi: 10.3897/zookeys.1089.78182. eCollection 2022. Zookeys. 2022. PMID: 35586600 Free PMC article.
-
Cicada Endosymbionts Have tRNAs That Are Correctly Processed Despite Having Genomes That Do Not Encode All of the tRNA Processing Machinery.mBio. 2019 Jun 18;10(3):e01950-18. doi: 10.1128/mBio.01950-18. mBio. 2019. PMID: 31213566 Free PMC article.
-
Comparative mitogenomics of Drosophilidae and the evolution of the Zygothrica genus group (Diptera, Drosophilidae).Genetica. 2021 Dec;149(5-6):267-281. doi: 10.1007/s10709-021-00132-8. Epub 2021 Oct 5. Genetica. 2021. PMID: 34609625
References
-
- Arita M., Suematsu T., Osanai A., Inaba T., Kamiya H., Kita K., et al. (2006). An evolutionary ‘intermediate state’ of mitochondrial translation systems found in Trichinella species of parasitic nematodes: co-evolution of tRNA and EF-Tu. Nucleic Acids Res. 34 5291–5929 10.1093/nar/gkl526 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

