Estimating coverage in metagenomic data sets and why it matters
- PMID: 24824669
- PMCID: PMC4992084
- DOI: 10.1038/ismej.2014.76
Estimating coverage in metagenomic data sets and why it matters
Figures
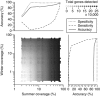
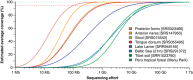
References
-
- Caro-Quintero A, Konstantinidis KT. (2012). Bacterial species may exist, metagenomics reveal. Environ Microbiol 14: 347–355. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

