Transcriptional regulation of oncogenic protein kinase Cϵ (PKCϵ) by STAT1 and Sp1 proteins
- PMID: 24825907
- PMCID: PMC4094091
- DOI: 10.1074/jbc.M114.548446
Transcriptional regulation of oncogenic protein kinase Cϵ (PKCϵ) by STAT1 and Sp1 proteins
Abstract
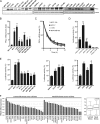
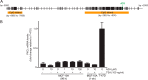
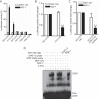
Overexpression of PKCϵ, a kinase associated with tumor aggressiveness and widely implicated in malignant transformation and metastasis, is a hallmark of multiple cancers, including mammary, prostate, and lung cancer. To characterize the mechanisms that control PKCϵ expression and its up-regulation in cancer, we cloned an ∼ 1.6-kb promoter segment of the human PKCϵ gene (PRKCE) that displays elevated transcriptional activity in cancer cells. A comprehensive deletional analysis established two regions rich in Sp1 and STAT1 sites located between -777 and -105 bp (region A) and -921 and -796 bp (region B), respectively, as responsible for the high transcriptional activity observed in cancer cells. A more detailed mutagenesis analysis followed by EMSA and ChIP identified Sp1 sites in positions -668/-659 and -269/-247 as well as STAT1 sites in positions -880/-869 and -793/-782 as the elements responsible for elevated promoter activity in breast cancer cells relative to normal mammary epithelial cells. RNAi silencing of Sp1 and STAT1 in breast cancer cells reduced PKCϵ mRNA and protein expression, as well as PRKCE promoter activity. Moreover, a strong correlation was found between PKCϵ and phospho-Ser-727 (active) STAT1 levels in breast cancer cells. Our results may have significant implications for the development of approaches to target PKCϵ and its effectors in cancer therapeutics.
Keywords: Breast Cancer; Diacylglycerol; Gene Expression; Protein Kinase; Protein Kinase C (PKCϵ); STAT Transcription Factor; Signal Transduction; Specificity Protein 1 (Sp1); Transcription Promoter.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Kampfer S., Windegger M., Hochholdinger F., Schwaiger W., Pestell R. G., Baier G., Grunicke H. H., Uberall F. (2001) Protein kinase C isoforms involved in the transcriptional activation of cyclin D1 by transforming Ha-Ras. J. Biol. Chem. 276, 42834–42842 - PubMed
-
- Mesquita R. F., Paul M. A., Valmaseda A., Francois A., Jabr R., Anjum S., Marber M. S., Budhram-Mahadeo V., Heads R. J. (2014) Protein kinase Cϵ-calcineurin cosignaling downstream of toll-like receptor 4 downregulates fibrosis and induces wound healing gene expression in cardiac myofibroblasts. Mol. Cell Biol. 34, 574–594 - PMC - PubMed
-
- Saurin A. T., Durgan J., Cameron A. J., Faisal A., Marber M. S., Parker P. J. (2008) The regulated assembly of a PKCϵ complex controls the completion of cytokinesis. Nat. Cell Biol. 10, 891–901 - PubMed
-
- Soh J. W., Weinstein I. B. (2003) Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J. Biol. Chem. 278, 34709–34716 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

