Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors
- PMID: 24828498
- PMCID: PMC4067223
- DOI: 10.1074/jbc.M114.564534
Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors
Abstract
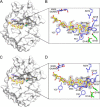
Small molecule inhibitors against chitinases have potential applications as pesticides, fungicides, and antiasthmatics. Here, we report that a series of fully deacetylated chitooligosaccharides (GlcN)2-7 can act as inhibitors against the insect chitinase OfChtI, the human chitinase HsCht, and the bacterial chitinases SmChiA and SmChiB with IC50 values at micromolar to millimolar levels. The injection of mixed (GlcN)2-7 into the fifth instar larvae of the insect Ostrinia furnacalis resulted in 85% of the larvae being arrested at the larval stage and death after 10 days, also suggesting that (GlcN)2-7 might inhibit OfChtI in vivo. Crystal structures of the catalytic domain of OfChtI (OfChtI-CAD) complexed with (GlcN)5,6 were obtained at resolutions of 2.0 Å. These structures, together with mutagenesis and thermodynamic analysis, suggested that the inhibition was strongly related to the interaction between the -1 GlcN residue of the inhibitor and the catalytic Glu(148) of the enzyme. Structure-based comparison showed that the fully deacetylated chitooligosaccharides mimic the substrate chitooligosaccharides by binding to the active cleft. This work first reports the inhibitory activity and proposed inhibitory mechanism of fully deacetylated chitooligosaccharides. Because the fully deacetylated chitooligosaccharides can be easily derived from chitin, one of the most abundant materials in nature, this work also provides a platform for developing eco-friendly inhibitors against chitinases.
Keywords: Chitinase; Crystal Structure; Deacetylated Chitooligosaccharides; Enzyme Inhibitor; Glycoside Hydrolase; Insect Molting; Thermodynamics.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Vaaje-Kolstad G., Horn S. J., Sørlie M., Eijsink V. G. (2013) The chitinolytic machinery of Serratia marcescens: a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 280, 3028–3049 - PubMed
-
- Genta F. A., Blanes L., Cristofoletti P. T., do Lago C. L., Terra W. R., Ferreira C. (2006) Purification, characterization and molecular cloning of the major chitinase from Tenebrio molitor larval midgut. Insect Biochem. Mol. Biol. 36, 789–800 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

