Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition
- PMID: 24829334
- PMCID: PMC4097766
- DOI: 10.1128/JVI.00737-14
Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition
Abstract
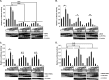
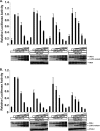
The interferon antiviral system is a primary barrier to virus replication triggered upon recognition of nonself RNAs by the cytoplasmic sensors encoded by retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology gene 2 (LGP2). Paramyxovirus V proteins are interferon antagonists that can selectively interact with MDA5 and LGP2 through contact with a discrete helicase domain region. Interaction with MDA5, an activator of antiviral signaling, disrupts interferon gene expression and antiviral responses. LGP2 has more diverse reported roles as both a coactivator of MDA5 and a negative regulator of both RIG-I and MDA5. This functional dichotomy, along with the concurrent interference with both cellular targets, has made it difficult to assess the unique consequences of V protein interaction with LGP2. To directly evaluate the impact of LGP2 interference, MDA5 and LGP2 variants unable to be recognized by measles virus and parainfluenza virus 5 (PIV5) V proteins were tested in signaling assays. Results indicate that interaction with LGP2 specifically prevents coactivation of MDA5 signaling and that LGP2's negative regulatory capacity was not affected. V proteins only partially antagonize RIG-I at high concentrations, and their expression had no additive effects on LGP2-mediated negative regulation. However, conversion of RIG-I to a direct V protein target was accomplished by only two amino acid substitutions that allowed both V protein interaction and efficient interference. These results clarify the unique consequences of MDA5 and LGP2 interference by paramyxovirus V proteins and help resolve the distinct roles of LGP2 in both activation and inhibition of antiviral signal transduction. Importance: Paramyxovirus V proteins interact with two innate immune receptors, MDA5 and LGP2, but not RIG-I. V proteins prevent MDA5 from signaling to the beta interferon promoter, but the consequences of LGP2 targeting are poorly understood. As the V protein targets MDA5 and LGP2 simultaneously, and LGP2 is both a positive and negative regulator of both MDA5 and RIG-I, it has been difficult to evaluate the specific advantages conferred by LGP2 targeting. Experiments with V-insensitive proteins revealed that the primary outcome of LGP2 interference is suppression of its ability to synergize with MDA5. LGP2's negative regulation of MDA5 and RIG-I remains intact irrespective of V protein interaction. Complementary experiments demonstrate that RIG-I can be converted to V protein sensitivity by two amino acid substitutions. These findings clarify the functions of LGP2 as a positive regulator of MDA5 signaling, demonstrate the basis for V-mediated LGP2 targeting, and broaden our understanding of paramyxovirus-host interactions.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2.J Virol. 2009 Jul;83(14):7252-60. doi: 10.1128/JVI.00153-09. Epub 2009 Apr 29. J Virol. 2009. PMID: 19403670 Free PMC article.
-
Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I.J Virol. 2013 Mar;87(5):2974-8. doi: 10.1128/JVI.02843-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269789 Free PMC article.
-
LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses.Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1512-7. doi: 10.1073/pnas.0912986107. Epub 2010 Jan 8. Proc Natl Acad Sci U S A. 2010. PMID: 20080593 Free PMC article.
-
LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review.
-
MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction.J Virol. 2014 Aug;88(15):8194-200. doi: 10.1128/JVI.00640-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850739 Free PMC article. Review.
Cited by
-
Chicken-Derived Pattern Recognition Receptor chLGP2 Inhibits the Replication and Proliferation of Infectious Bronchitis Virus.Front Microbiol. 2022 Jan 25;12:810215. doi: 10.3389/fmicb.2021.810215. eCollection 2021. Front Microbiol. 2022. PMID: 35145497 Free PMC article.
-
Antiviral RNA recognition and assembly by RLR family innate immune sensors.Cytokine Growth Factor Rev. 2014 Oct;25(5):507-12. doi: 10.1016/j.cytogfr.2014.07.006. Epub 2014 Jul 15. Cytokine Growth Factor Rev. 2014. PMID: 25081315 Free PMC article. Review.
-
Viral Respiratory Pathogens and Lung Injury.Clin Microbiol Rev. 2021 Mar 31;34(3):e00103-20. doi: 10.1128/CMR.00103-20. Print 2021 Jun 16. Clin Microbiol Rev. 2021. PMID: 33789928 Free PMC article. Review.
-
RIG-I in RNA virus recognition.Virology. 2015 May;479-480:110-21. doi: 10.1016/j.virol.2015.02.017. Epub 2015 Mar 5. Virology. 2015. PMID: 25749629 Free PMC article. Review.
-
LGP2 Promotes Type I Interferon Production To Inhibit PRRSV Infection via Enhancing MDA5-Mediated Signaling.J Virol. 2023 Jan 31;97(1):e0184322. doi: 10.1128/jvi.01843-22. Epub 2023 Jan 9. J Virol. 2023. PMID: 36622220 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

