Emergence of CD4 independence envelopes and astrocyte infection in R5 simian-human immunodeficiency virus model of encephalitis
- PMID: 24829360
- PMCID: PMC4135954
- DOI: 10.1128/JVI.01237-14
Emergence of CD4 independence envelopes and astrocyte infection in R5 simian-human immunodeficiency virus model of encephalitis
Abstract
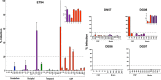
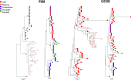
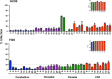
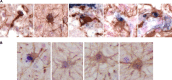
Human immunodeficiency virus type 1 (HIV-1) infection in the central nervous system (CNS) is characterized by replication in macrophages or brain microglia that express low levels of the CD4 receptor and is the cause of HIV-associated dementia and related cognitive and motor disorders that affect 20 to 30% of treatment-naive patients with AIDS. Independent viral envelope evolution in the brain has been reported, with the need for robust replication in resident CD4(low) cells, as well as CD4-negative cells, such as astrocytes, proposed as a major selective pressure. We previously reported giant-cell encephalitis in subtype B and C R5 simian-human immunodeficiency virus (SHIV)-infected macaques (SHIV-induced encephalitis [SHIVE]) that experienced very high chronic viral loads and progressed rapidly to AIDS, with varying degrees of macrophage or microglia infection and activation of these immune cells, as well as astrocytes, in the CNS. In this study, we characterized envelopes (Env) amplified from the brains of subtype B and C R5 SHIVE macaques. We obtained data in support of an association between severe neuropathological changes, robust macrophage and microglia infection, and evolution to CD4 independence. Moreover, the degree of Env CD4 independence appeared to correlate with the extent of astrocyte infection in vivo. These findings further our knowledge of the CNS viral population phenotypes that are associated with the severity of HIV/SHIV-induced neurological injury and improve our understanding of the mechanism of HIV-1 cellular tropism and persistence in the brain.
Importance: Human immunodeficiency virus type 1 (HIV-1) infection of astrocytes in the brain has been suggested to be important in HIV persistence and neuropathogenesis but has not been definitively demonstrated in an animal model of HIV-induced encephalitis (HIVE). Here, we describe a new nonhuman primate (NHP) model of R5 simian-human immunodeficiency virus (SHIV)-induced encephalitis (SHIVE) with several classical HIVE features that include astrocyte infection. We further show an association between severe neuropathological changes, robust resident microglia infection, and evolution to CD4 independence of viruses in the central nervous system (CNS), with expansion to infection of truly CD4-negative cells in vivo. These findings support the use of the R5 SHIVE models to study the contribution of the HIV envelope and viral clades to neurovirulence and residual virus replication in the CNS, providing information that should guide efforts to eradicate HIV from the body.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Simian Immunodeficiency Virus-Infected Memory CD4+ T Cells Infiltrate to the Site of Infected Macrophages in the Neuroparenchyma of a Chronic Macaque Model of Neurological Complications of AIDS.mBio. 2020 Apr 21;11(2):e00602-20. doi: 10.1128/mBio.00602-20. mBio. 2020. PMID: 32317323 Free PMC article.
-
Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies.J Neurovirol. 2008 Aug;14(4):318-26. doi: 10.1080/13550280802132857. J Neurovirol. 2008. PMID: 18780233 Free PMC article. Review.
-
Simian-human immunodeficiency virus containing a human immunodeficiency virus type 1 subtype-E envelope gene: persistent infection, CD4(+) T-cell depletion, and mucosal membrane transmission in macaques.J Virol. 2000 Sep;74(17):7851-60. doi: 10.1128/jvi.74.17.7851-7860.2000. J Virol. 2000. PMID: 10933692 Free PMC article.
-
Central Nervous System Inflammation and Infection during Early, Nonaccelerated Simian-Human Immunodeficiency Virus Infection in Rhesus Macaques.J Virol. 2018 May 14;92(11):e00222-18. doi: 10.1128/JVI.00222-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29563297 Free PMC article.
-
The SIV Envelope Glycoprotein, Viral Tropism, and Pathogenesis: Novel Insights from Nonhuman Primate Models of AIDS.Curr HIV Res. 2018;16(1):29-40. doi: 10.2174/1570162X15666171124123116. Curr HIV Res. 2018. PMID: 29173176 Review.
Cited by
-
Transmitted/founder SHIV.D replicates in the brain, causes neuropathogenesis, and persists on combination antiretroviral therapy in rhesus macaques.Retrovirology. 2023 Aug 10;20(1):13. doi: 10.1186/s12977-023-00628-5. Retrovirology. 2023. PMID: 37563642 Free PMC article.
-
Gp120 V5 Is Targeted by the First Wave of Sequential Neutralizing Antibodies in SHIVSF162P3N-Infected Rhesus Macaques.Viruses. 2018 May 16;10(5):262. doi: 10.3390/v10050262. Viruses. 2018. PMID: 29772652 Free PMC article.
-
Productive HIV infection in astrocytes can be established via a nonclassical mechanism.AIDS. 2020 Jun 1;34(7):963-978. doi: 10.1097/QAD.0000000000002512. AIDS. 2020. PMID: 32379159 Free PMC article.
-
HIV-1 endocytosis in astrocytes: a kiss of death or survival of the fittest?Neurosci Res. 2014 Nov;88:16-22. doi: 10.1016/j.neures.2014.08.013. Epub 2014 Sep 8. Neurosci Res. 2014. PMID: 25219546 Free PMC article. Review.
-
Astrocytes are HIV reservoirs in the brain: A cell type with poor HIV infectivity and replication but efficient cell-to-cell viral transfer.J Neurochem. 2021 Jul;158(2):429-443. doi: 10.1111/jnc.15336. Epub 2021 Mar 22. J Neurochem. 2021. PMID: 33655498 Free PMC article.
References
-
- Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, CHARTER Group 2010. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75:2087–2096. 10.1212/WNL.0b013e318200d727 - DOI - PMC - PubMed
-
- Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner JP, Tawadros R, Pomerantz RJ. 1996. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS 10:573–585. 10.1097/00002030-199606000-00002 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

