Dynactin integrity depends upon direct binding of dynamitin to Arp1
- PMID: 24829381
- PMCID: PMC4091830
- DOI: 10.1091/mbc.E14-03-0842
Dynactin integrity depends upon direct binding of dynamitin to Arp1
Abstract
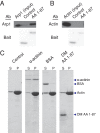
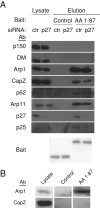
Dynactin is a multiprotein complex that works with cytoplasmic dynein and other motors to support a wide range of cell functions. It serves as an adaptor that binds both dynein and cargoes and enhances single-motor processivity. The dynactin subunit dynamitin (also known as p50) is believed to be integral to dynactin structure because free dynamitin displaces the dynein-binding p150(Glued) subunit from the cargo-binding Arp1 filament. We show here that the intrinsically disordered dynamitin N-terminus binds to Arp1 directly. When expressed in cells, dynamitin amino acids (AA) 1-87 causes complete release of endogenous dynamitin, p150, and p24 from dynactin, leaving behind Arp1 filaments carrying the remaining dynactin subunits (CapZ, p62, Arp11, p27, and p25). Tandem-affinity purification-tagged dynamitin AA 1-87 binds the Arp filament specifically, and binding studies with purified native Arp1 reveal that this fragment binds Arp1 directly. Neither CapZ nor the p27/p25 dimer contributes to interactions between dynamitin and the Arp filament. This work demonstrates for the first time that Arp1 can directly bind any protein besides another Arp and provides important new insight into the underpinnings of dynactin structure.
© 2014 Cheong et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures




References
-
- Bingham JB, Schroer TA. Self-regulated polymerization of the actin-related protein. Arp1 Curr Biol. 1999;9:223–226. - PubMed
-
- Brown CL, Maier KC, Stauber T, Ginkel LM, Wordeman L, Vernos I, Schroer TA. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 2005;6:1114–1124. - PubMed
-
- Chen YH, Yang JT. A new approach to the calculation of secondary structures of globular proteins by optical rotatory dispersion and circular dichroism. Biochem Biophys Res Commun. 1971;44:1285–1291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

