PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes
- PMID: 24829450
- PMCID: PMC4086120
- DOI: 10.1093/nar/gku397
PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes
Abstract
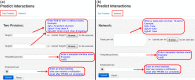
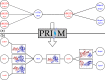
The PRISM web server enables fast and accurate prediction of protein-protein interactions (PPIs). The prediction algorithm is knowledge-based. It combines structural similarity and accounts for evolutionary conservation in the template interfaces. The predicted models are stored in its repository. Given two protein structures, PRISM will provide a structural model of their complex if a matching template interface is available. Users can download the complex structure, retrieve the interface residues and visualize the complex model. The PRISM web server is user friendly, free and open to all users at http://cosbi.ku.edu.tr/prism.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

