Hydrolytic function of Exo1 in mammalian mismatch repair
- PMID: 24829455
- PMCID: PMC4066806
- DOI: 10.1093/nar/gku420
Hydrolytic function of Exo1 in mammalian mismatch repair
Abstract
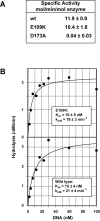
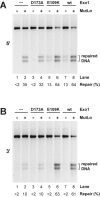
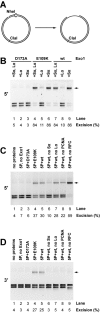
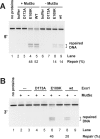
Genetic and biochemical studies have previously implicated exonuclease 1 (Exo1) in yeast and mammalian mismatch repair, with results suggesting that function of the protein in the reaction depends on both its hydrolytic activity and its ability to interact with other components of the repair system. However, recent analysis of an Exo1-E109K knockin mouse has concluded that Exo1 function in mammalian mismatch repair is restricted to a structural role, a conclusion based on a prior report that N-terminal His-tagged Exo1-E109K is hydrolytically defective. Because Glu-109 is distant from the nuclease hydrolytic center, we have compared the activity of untagged full-length Exo1-E109K with that of wild type Exo1 and the hydrolytically defective active site mutant Exo1-D173A. We show that the activity of Exo1-E109K is comparable to that of wild type enzyme in a conventional exonuclease assay and that in contrast to a D173A active site mutant, Exo1-E109K is fully functional in mismatch-provoked excision and repair. We conclude that the catalytic function of Exo1 is required for its participation in mismatch repair. We also consider the other phenotypes of the Exo1-E109K mouse in the context of Exo1 hydrolytic function.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Tran P.T., Erdeniz N., Symington L.S., Liskay R.M. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559.. - PubMed
-
- Szankasi P., Smith G.R. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 1992;267:3014–3023.. - PubMed
-
- Szankasi P., Smith G.R. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169.. - PubMed
-
- Rudolph C., Fleck O., Kohli J. Schizosaccharomyces pombe exo1 is involved in the same mismatch repair pathway as msh2 and pms1. Curr. Genet. 1998;34:343–350.. - PubMed
-
- Dherin C., Gueneau E., Francin M., Nunez M., Miron S., Liberti S.E., Rasmussen L.J., Zinn-Justin S., Gilquin B., Charbonnier J.B., et al. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 2009;29:907–918.. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

