Accelerated receptor shedding inhibits kidney injury molecule-1 (KIM-1)-mediated efferocytosis
- PMID: 24829508
- PMCID: PMC4380482
- DOI: 10.1152/ajprenal.00638.2013
Accelerated receptor shedding inhibits kidney injury molecule-1 (KIM-1)-mediated efferocytosis
Abstract
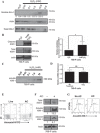
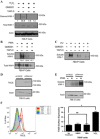
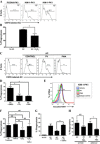
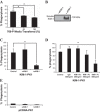
Efficient clearance of apoptotic cells (efferocytosis) prevents inflammation and permits repair following tissue injury. Kidney injury molecule-1 (KIM-1) is a receptor for phosphatidylserine, an "eat-me" signal exposed on the surface of apoptotic cells that marks them for phagocytic clearance. KIM-1 is upregulated on proximal tubule epithelial cells (PTECs) during ischemic acute kidney injury (AKI), enabling efferocytosis by surviving PTECs. KIM-1 is spontaneously cleaved at its ectodomain region to generate a soluble fragment that serves a sensitive and specific biomarker for AKI, but the biological relevance of KIM-1 shedding is unknown. Here, we sought to determine how KIM-1 shedding might regulate efferocytosis. Using cells that endogenously and exogenously express KIM-1, we found that hydrogen peroxide-mediated oxidative injury or PMA treatment accelerated KIM-1 shedding in a dose-dependent manner. KIM-1 shedding was also accelerated when apoptotic cells were added. Accelerated shedding or the presence of excess soluble KIM-1 in the extracellular milieu significantly inhibited efferocytosis. We also identified that TNF-α-converting enzyme (TACE or ADAM17) mediates both the spontaneous and PMA-accelerated shedding of KIM-1. While accelerated shedding inhibited efferocytosis, we found that spontaneous KIM-1 cleavage does not affect the phagocytic efficiency of PTECs. Our results suggest that KIM-1 shedding is accelerated by worsening cellular injury, and excess soluble KIM-1 competitively inhibits efferocytosis. These findings may be important in AKI when there is severe cellular injury.
Keywords: efferocytosis; kidney injury molecule-1 (KIM-1); proximal tubule epithelial cells (PTECs); receptor; shedding.
Copyright © 2014 the American Physiological Society.
Figures


Similar articles
-
Mapping and functional characterization of murine kidney injury molecule-1 proteolytic cleavage site.Mol Cell Biochem. 2021 Feb;476(2):1093-1108. doi: 10.1007/s11010-020-03975-5. Epub 2020 Nov 19. Mol Cell Biochem. 2021. PMID: 33211259
-
Tctex-1, a novel interaction partner of Kidney Injury Molecule-1, is required for efferocytosis.J Cell Physiol. 2018 Oct;233(10):6877-6895. doi: 10.1002/jcp.26578. Epub 2018 Apr 25. J Cell Physiol. 2018. PMID: 29693725 Free PMC article.
-
KIM-1-mediated anti-inflammatory activity is preserved by MUC1 induction in the proximal tubule during ischemia-reperfusion injury.Am J Physiol Renal Physiol. 2021 Aug 1;321(2):F135-F148. doi: 10.1152/ajprenal.00127.2021. Epub 2021 Jun 21. Am J Physiol Renal Physiol. 2021. PMID: 34151589 Free PMC article.
-
Kidney injury molecule-1: a translational journey.Trans Am Clin Climatol Assoc. 2014;125:293-9; discussion 299. Trans Am Clin Climatol Assoc. 2014. PMID: 25125746 Free PMC article. Review.
-
Kidney injury molecule-1.Curr Opin Crit Care. 2010 Dec;16(6):556-61. doi: 10.1097/MCC.0b013e32834008d3. Curr Opin Crit Care. 2010. PMID: 20930626 Review.
Cited by
-
The cisplatin-induced acute kidney injury is a novel risk factor for postoperative complications in patients with esophageal cancer: a retrospective cohort study.BMC Surg. 2023 Mar 27;23(1):67. doi: 10.1186/s12893-023-01949-0. BMC Surg. 2023. PMID: 36973771 Free PMC article.
-
New Answers to Old Conundrums: What Antibodies, Exosomes and Inflammasomes Bring to the Conversation. Canadian National Transplant Research Program International Summit Report.Transplantation. 2018 Feb;102(2):209-214. doi: 10.1097/TP.0000000000001872. Transplantation. 2018. PMID: 28731910 Free PMC article. Review.
-
ADAM10 and ADAM17, Major Regulators of Chronic Kidney Disease Induced Atherosclerosis?Int J Mol Sci. 2023 Apr 15;24(8):7309. doi: 10.3390/ijms24087309. Int J Mol Sci. 2023. PMID: 37108478 Free PMC article. Review.
-
Translational Assessment of Drug-Induced Proximal Tubule Injury Using a Kidney Microphysiological System.CPT Pharmacometrics Syst Pharmacol. 2019 May;8(5):316-325. doi: 10.1002/psp4.12400. Epub 2019 Apr 9. CPT Pharmacometrics Syst Pharmacol. 2019. PMID: 30869201 Free PMC article.
-
ADAMs family in kidney physiology and pathology.EBioMedicine. 2021 Oct;72:103628. doi: 10.1016/j.ebiom.2021.103628. Epub 2021 Oct 12. EBioMedicine. 2021. PMID: 34653870 Free PMC article. Review.
References
-
- Anderson HA, Maylock CA, Williams JA, Paweletz CP, Shu H, Shacter E. Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat Immunol 4: 87–91, 2003. - PubMed
-
- Arribas J, Coodly L, Vollmer P, Kishimoto TK, Rose-John S, Massague J. Diverse cell surface protein ectodomains are shed by a system sensitive to metalloprotease inhibitors. J Biol Chem 271: 11376–11382, 1996. - PubMed
-
- Arribas J, Esselens C. ADAM17 as a therapeutic target in multiple diseases. Curr Pharm Des 15: 2319–2335, 2009. - PubMed
-
- Arroyo A, Modriansky M, Serinkan FB, Bello RI, Matsura T, Jiang J, Tyurin VA, Tyurina YY, Fadeel B, Kagan VE. NADPH oxidase-dependent oxidation and externalization of phosphatidylserine during apoptosis in Me2SO-differentiated HL-60 cells. Role in phagocytic clearance. J Biol Chem 277: 49965–49975, 2002. - PubMed
-
- Atabai K, Jame S, Azhar N, Kuo A, Lam M, McKleroy W, Dehart G, Rahman S, Xia DD, Melton AC, Wolters P, Emson CL, Turner SM, Werb Z, Sheppard D. Mfge8 diminishes the severity of tissue fibrosis in mice by binding and targeting collagen for uptake by macrophages. J Clin Invest 119: 3713–3722, 2009. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

