Genetic Control and Evolution of Anthocyanin Methylation
- PMID: 24830298
- PMCID: PMC4081349
- DOI: 10.1104/pp.113.234526
Genetic Control and Evolution of Anthocyanin Methylation
Abstract
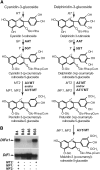
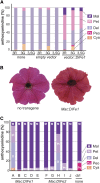
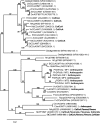
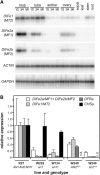
Anthocyanins are a chemically diverse class of secondary metabolites that color most flowers and fruits. They consist of three aromatic rings that can be substituted with hydroxyl, sugar, acyl, and methyl groups in a variety of patterns depending on the plant species. To understand how such chemical diversity evolved, we isolated and characterized METHYLATION AT THREE2 (MT2) and the two METHYLATION AT FIVE (MF) loci from Petunia spp., which direct anthocyanin methylation in petals. The proteins encoded by MT2 and the duplicated MF1 and MF2 genes and a putative grape (Vitis vinifera) homolog Anthocyanin O-Methyltransferase1 (VvAOMT1) are highly similar to and apparently evolved from caffeoyl-Coenzyme A O-methyltransferases by relatively small alterations in the active site. Transgenic experiments showed that the Petunia spp. and grape enzymes have remarkably different substrate specificities, which explains part of the structural anthocyanin diversity in both species. Most strikingly, VvAOMT1 expression resulted in the accumulation of novel anthocyanins that are normally not found in Petunia spp., revealing how alterations in the last reaction can reshuffle the pathway and affect (normally) preceding decoration steps in an unanticipated way. Our data show how variations in gene expression patterns, loss-of-function mutations, and alterations in substrate specificities all contributed to the anthocyanins' structural diversity.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures




Similar articles
-
CCoAOMT Down-Regulation Activates Anthocyanin Biosynthesis in Petunia.Plant Physiol. 2016 Feb;170(2):717-31. doi: 10.1104/pp.15.01646. Epub 2015 Nov 30. Plant Physiol. 2016. PMID: 26620524 Free PMC article.
-
On the origin of anthocyanin methyltransferase isozymes of Petunia hybrida and their role in regulation of anthocyanin methylation.Theor Appl Genet. 1984 Aug;68(5):459-66. doi: 10.1007/BF00254821. Theor Appl Genet. 1984. PMID: 24257738
-
CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production.Microb Cell Fact. 2017 Jan 17;16(1):10. doi: 10.1186/s12934-016-0623-3. Microb Cell Fact. 2017. PMID: 28095853 Free PMC article.
-
Properties and genetic control of four methyltransferases involved in methylation of anthocyanins in flowers of Petunia hybrida.Planta. 1984 Feb;160(2):174-9. doi: 10.1007/BF00392867. Planta. 1984. PMID: 24258421
-
Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration.Plant Cell Physiol. 2015 Jan;56(1):28-40. doi: 10.1093/pcp/pcu097. Epub 2014 Jul 10. Plant Cell Physiol. 2015. PMID: 25015943 Review.
Cited by
-
Diversification of Chemical Structures of Methoxylated Flavonoids and Genes Encoding Flavonoid-O-Methyltransferases.Plants (Basel). 2022 Feb 21;11(4):564. doi: 10.3390/plants11040564. Plants (Basel). 2022. PMID: 35214897 Free PMC article. Review.
-
Establishment of an efficient callus transient transformation system for Vitis vinifera cv. 'Chardonnay'.Protoplasma. 2024 Mar;261(2):351-366. doi: 10.1007/s00709-023-01901-2. Epub 2023 Oct 31. Protoplasma. 2024. PMID: 37906315
-
Global organization of phenylpropanoid and anthocyanin pathways revealed by proximity labeling of trans-cinnamic acid 4-hydroxylase in Petunia inflata petal protoplasts.Front Plant Sci. 2024 Sep 19;15:1295750. doi: 10.3389/fpls.2024.1295750. eCollection 2024. Front Plant Sci. 2024. PMID: 39363925 Free PMC article.
-
Evolutionary walks through flower colour space driven by gene expression in Petunia and allies (Petunieae).Proc Biol Sci. 2023 Jul 12;290(2002):20230275. doi: 10.1098/rspb.2023.0275. Epub 2023 Jul 5. Proc Biol Sci. 2023. PMID: 37403504 Free PMC article.
-
CCoAOMT Down-Regulation Activates Anthocyanin Biosynthesis in Petunia.Plant Physiol. 2016 Feb;170(2):717-31. doi: 10.1104/pp.15.01646. Epub 2015 Nov 30. Plant Physiol. 2016. PMID: 26620524 Free PMC article.
References
-
- Acevedo De la Cruz A, Hilbert G, Rivière C, Mengin V, Ollat N, Bordenave L, Decroocq S, Delaunay JC, Delrot S, Mérillon JM, et al. (2012) Anthocyanin identification and composition of wild Vitis spp. accessions by using LC-MS and LC-NMR. Anal Chim Acta 732: 145–152 - PubMed
-
- Akita Y, Kitamura S, Hase Y, Narumi I, Ishizaka H, Kondo E, Kameari N, Nakayama M, Tanikawa N, Morita Y, et al. (2011) Isolation and characterization of the fragrant cyclamen O-methyltransferase involved in flower coloration. Planta 234: 1127–1136 - PubMed
-
- Ando T, Saito N, Tatsuzawa F, Kakefuda T, Yamakage K, Ohtani E, Koshi-ishi M, Matsusake Y, Kokubun H, Watanabe H, et al. (1999) Floral anthocyanins in wild taxa of Petunia (Solanaceae). Biochem Syst Ecol 27: 623–650
-
- Brouillard R, Chassaing S, Fougerousse A. (2003) Why are grape/fresh wine anthocyanins so simple and why is it that red wine color lasts so long? Phytochemistry 64: 1179–1186 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

