A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum
- PMID: 24830312
- PMCID: PMC4022464
- DOI: 10.1371/journal.pgen.1004382
A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum
Abstract
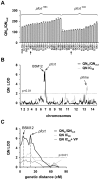
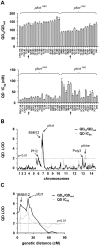
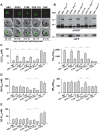
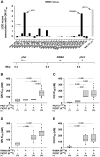
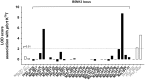
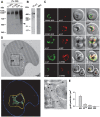
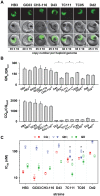
The emerging resistance to quinine jeopardizes the efficacy of a drug that has been used in the treatment of malaria for several centuries. To identify factors contributing to differential quinine responses in the human malaria parasite Plasmodium falciparum, we have conducted comparative quantitative trait locus analyses on the susceptibility to quinine and also its stereoisomer quinidine, and on the initial and steady-state intracellular drug accumulation levels in the F1 progeny of a genetic cross. These data, together with genetic screens of field isolates and laboratory strains associated differential quinine and quinidine responses with mutated pfcrt, a segment on chromosome 13, and a novel candidate gene, termed MAL7P1.19 (encoding a HECT ubiquitin ligase). Despite a strong likelihood of association, episomal transfections demonstrated a role for the HECT ubiquitin-protein ligase in quinine and quinidine sensitivity in only a subset of genetic backgrounds, and here the changes in IC50 values were moderate (approximately 2-fold). These data show that quinine responsiveness is a complex genetic trait with multiple alleles playing a role and that more experiments are needed to unravel the role of the contributing factors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
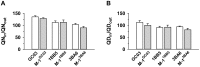

Similar articles
-
In vitro evaluation of quinidine sensitivity in Brazilian Plasmodium falciparum isolates: comparative analysis to quinine and chloroquine.Rev Inst Med Trop Sao Paulo. 2001 Jul-Aug;43(4):221-6. doi: 10.1590/s0036-46652001000400009. Rev Inst Med Trop Sao Paulo. 2001. PMID: 11558003
-
The in vitro response of Plasmodium falciparum to amodiaquine, quinine and quinidine in Tanga region, Tanzania.East Afr Med J. 1990 May;67(5):336-40. East Afr Med J. 1990. PMID: 2202587
-
In vivo and in vitro responses to quinine and quinidine of Plasmodium falciparum.Bull World Health Organ. 1988;66(3):347-52. Bull World Health Organ. 1988. PMID: 3048758 Free PMC article.
-
Update on genetic markers of quinine resistance in Plasmodium falciparum.Mol Biochem Parasitol. 2011 Jun;177(2):77-82. doi: 10.1016/j.molbiopara.2011.01.012. Epub 2011 Feb 1. Mol Biochem Parasitol. 2011. PMID: 21295079 Review.
-
Emerging drug--resistance and guidelines for treatment of malaria.J Coll Physicians Surg Pak. 2004 May;14(5):319-24. J Coll Physicians Surg Pak. 2004. PMID: 15225468 Review.
Cited by
-
pH-dependence of the Plasmodium falciparum chloroquine resistance transporter is linked to the transport cycle.Nat Commun. 2023 Jul 15;14(1):4234. doi: 10.1038/s41467-023-39969-2. Nat Commun. 2023. PMID: 37454114 Free PMC article.
-
Overexpression in Plasmodium falciparum of an intrinsically disordered protein segment of PfUT impairs the parasite's proteostasis and reduces its growth rate.Front Cell Infect Microbiol. 2025 May 13;15:1565814. doi: 10.3389/fcimb.2025.1565814. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40433665 Free PMC article.
-
A Plasmodium falciparum RING Finger E3 Ubiquitin Ligase Modifies the Roles of PfMDR1 and PfCRT in Parasite Drug Responses.Antimicrob Agents Chemother. 2023 Feb 16;67(2):e0082122. doi: 10.1128/aac.00821-22. Epub 2023 Jan 10. Antimicrob Agents Chemother. 2023. PMID: 36625569 Free PMC article.
-
Plasmodium Genomics and Genetics: New Insights into Malaria Pathogenesis, Drug Resistance, Epidemiology, and Evolution.Clin Microbiol Rev. 2019 Jul 31;32(4):e00019-19. doi: 10.1128/CMR.00019-19. Print 2019 Sep 18. Clin Microbiol Rev. 2019. PMID: 31366610 Free PMC article. Review.
-
Genomic and Genetic Approaches to Studying Antimalarial Drug Resistance and Plasmodium Biology.Trends Parasitol. 2021 Jun;37(6):476-492. doi: 10.1016/j.pt.2021.02.007. Epub 2021 Mar 11. Trends Parasitol. 2021. PMID: 33715941 Free PMC article. Review.
References
-
- World Health Organization (2012) World Malaria Report 2012. Geneva: World Health Organization.
-
- World Health Organization (2010) Guidelines for the treatment of malaria — 2nd edition. Geneva: World Health Organization.
-
- Pukrittayakamee S, Supanaranond W, Looareesuwan S, Vanijanonta S, White NJ (1994) Quinine in severe falciparum malaria: evidence of declining efficacy in Thailand. Trans R Soc Trop Med Hyg 88: 324–327. - PubMed
-
- Huttinger F, Satimai W, Wernsdorfer G, Wiedermann U, Congpuong K, et al. (2010) Sensitivity to artemisinin, mefloquine and quinine of Plasmodium falciparum in northwestern Thailand. Wien Klin Wochenschr 122 Suppl 352–56. - PubMed
-
- Chaijaroenkul W, Wisedpanichkij R, Na-Bangchang K (2010) Monitoring of in vitro susceptibilities and molecular markers of resistance of Plasmodium falciparum isolates from Thai-Myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop 113: 190–194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

